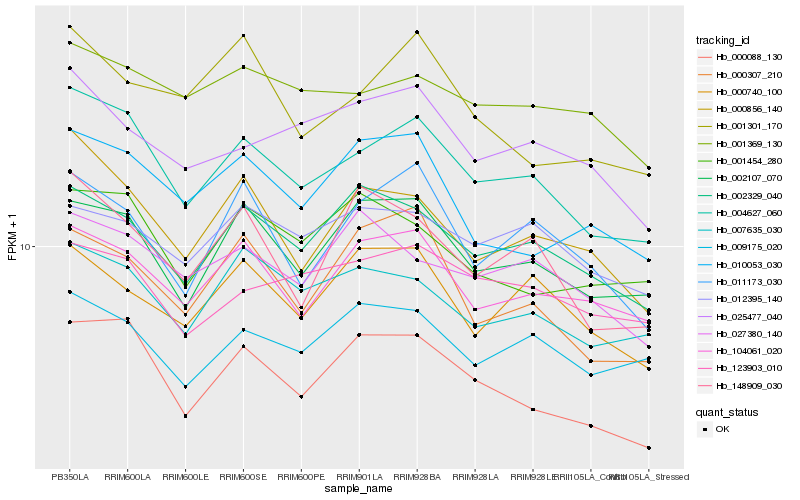
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004627_060 |
0.0 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 2 |
Hb_000088_130 |
0.0646342295 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 3 |
Hb_002107_070 |
0.0711534346 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 4 |
Hb_000856_140 |
0.0722510206 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 5 |
Hb_104061_020 |
0.0737018328 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 6 |
Hb_007635_030 |
0.0751711383 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_002329_040 |
0.0763680712 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 8 |
Hb_009175_020 |
0.0841048366 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_011173_030 |
0.0844526699 |
- |
- |
pumilio, putative [Ricinus communis] |
| 10 |
Hb_148909_030 |
0.0855716957 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001369_130 |
0.0862423365 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_025477_040 |
0.0866583152 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 13 |
Hb_123903_010 |
0.0876004812 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010053_030 |
0.0881401358 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 15 |
Hb_001454_280 |
0.0882147509 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 16 |
Hb_000307_210 |
0.0888782888 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000740_100 |
0.0896552481 |
- |
- |
calpain, putative [Ricinus communis] |
| 18 |
Hb_012395_140 |
0.0904556212 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 19 |
Hb_001301_170 |
0.0906839374 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 20 |
Hb_027380_140 |
0.090847847 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |