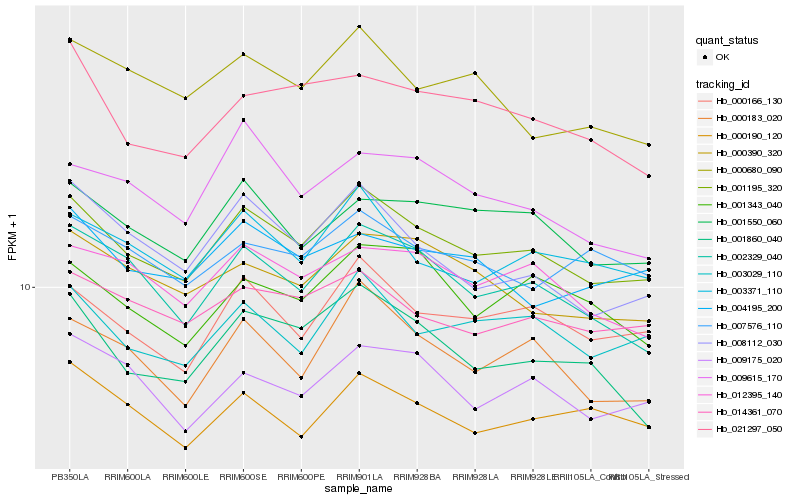
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_320 |
0.0 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 2 |
Hb_000166_130 |
0.0478140276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003371_110 |
0.0515174897 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 4 |
Hb_002329_040 |
0.052613225 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 5 |
Hb_003029_110 |
0.0536650669 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 6 |
Hb_014361_070 |
0.0554131713 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 7 |
Hb_000183_020 |
0.0559371865 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001550_060 |
0.0563311376 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 9 |
Hb_012395_140 |
0.0563402088 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 10 |
Hb_000390_320 |
0.0596085599 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 11 |
Hb_009175_020 |
0.0612097562 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_001860_040 |
0.0622611145 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 13 |
Hb_008112_030 |
0.0627027687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004195_200 |
0.0637170447 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 15 |
Hb_000190_120 |
0.0642886502 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 16 |
Hb_000680_090 |
0.0643935408 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 17 |
Hb_001343_040 |
0.064406705 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 18 |
Hb_021297_050 |
0.0648684413 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 19 |
Hb_007576_110 |
0.0653918389 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 20 |
Hb_009615_170 |
0.0665909287 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |