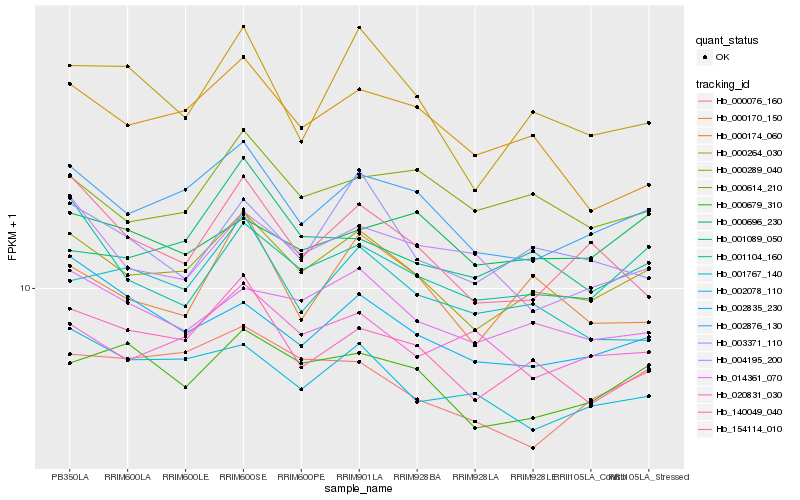
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000289_040 |
0.0 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 2 |
Hb_002876_130 |
0.039936671 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 3 |
Hb_140049_040 |
0.0510996601 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 4 |
Hb_014361_070 |
0.0521895092 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 5 |
Hb_000679_310 |
0.0558972735 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001104_160 |
0.0592830121 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 7 |
Hb_000614_210 |
0.0625197725 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003371_110 |
0.0636440487 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 9 |
Hb_000264_030 |
0.0638743987 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 10 |
Hb_000170_150 |
0.065256918 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002078_110 |
0.0671548378 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_002835_230 |
0.0683632848 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000076_160 |
0.0684240828 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 14 |
Hb_020831_030 |
0.0688851318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001089_050 |
0.0692495689 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_000174_060 |
0.0700927484 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 17 |
Hb_004195_200 |
0.070107652 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 18 |
Hb_000696_230 |
0.0701478582 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 19 |
Hb_001767_140 |
0.0704222495 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 20 |
Hb_154114_010 |
0.0704904258 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |