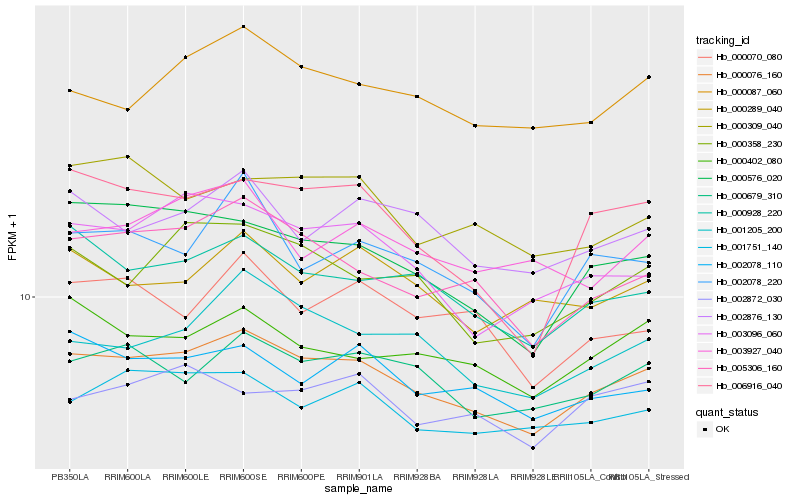
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_160 |
0.0 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 2 |
Hb_005306_160 |
0.0530046307 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_000928_220 |
0.0552670058 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 4 |
Hb_000679_310 |
0.0569535049 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000358_230 |
0.0595278364 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 6 |
Hb_000087_060 |
0.0603427433 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_001751_140 |
0.0605568827 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000576_020 |
0.061643544 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 9 |
Hb_002876_130 |
0.0628540286 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 10 |
Hb_001205_200 |
0.0629857415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002078_110 |
0.0634720369 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_000402_080 |
0.0636306964 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 13 |
Hb_003096_060 |
0.0642052429 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 14 |
Hb_000309_040 |
0.0671443731 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 15 |
Hb_000289_040 |
0.0684240828 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 16 |
Hb_002078_220 |
0.068976852 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006916_040 |
0.0697364028 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 18 |
Hb_003927_040 |
0.0701553723 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 19 |
Hb_000070_080 |
0.0705499616 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002872_030 |
0.0705866323 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |