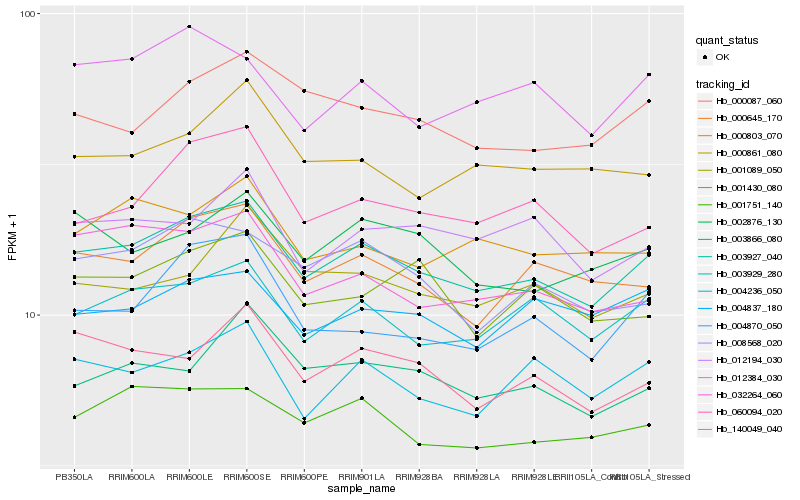
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003927_040 |
0.0 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 2 |
Hb_003929_280 |
0.0522026931 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 3 |
Hb_001751_140 |
0.0538427567 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000645_170 |
0.0548032278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_060094_020 |
0.0602960339 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 6 |
Hb_008568_020 |
0.0610039582 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 7 |
Hb_140049_040 |
0.0618111666 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 8 |
Hb_000087_060 |
0.0618581843 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 9 |
Hb_004236_050 |
0.0619485606 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 10 |
Hb_004870_050 |
0.0647287651 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 11 |
Hb_012384_030 |
0.0647567767 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 12 |
Hb_003866_080 |
0.0657189072 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 13 |
Hb_004837_180 |
0.0659279062 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002876_130 |
0.0659519578 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 15 |
Hb_032264_060 |
0.0665936922 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 16 |
Hb_000803_070 |
0.0668477088 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 17 |
Hb_001089_050 |
0.0671685118 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 18 |
Hb_000861_080 |
0.0679680038 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 19 |
Hb_012194_030 |
0.06816449 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001430_080 |
0.0685633525 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |