| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
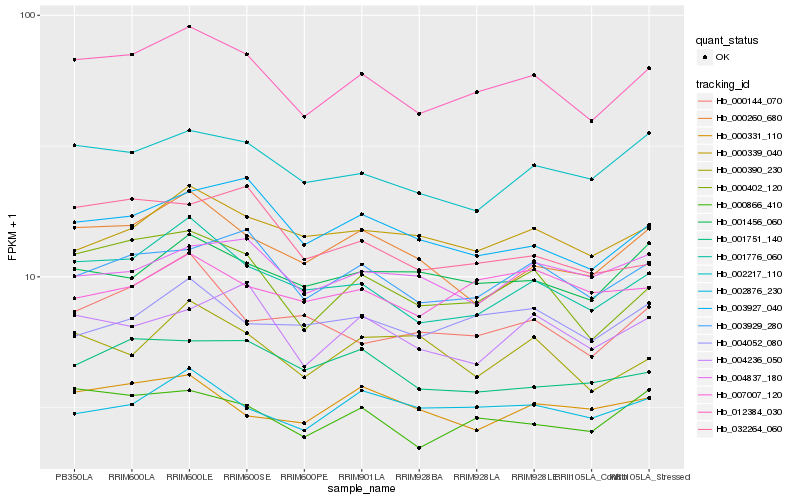
Hb_012384_030 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 2 |
Hb_000402_120 |
0.0476716506 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 3 |
Hb_001776_060 |
0.0489601359 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 4 |
Hb_000866_410 |
0.0570450595 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 5 |
Hb_003929_280 |
0.0587314647 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 6 |
Hb_002217_110 |
0.0633330304 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 7 |
Hb_003927_040 |
0.0647567767 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 8 |
Hb_000260_680 |
0.0676293201 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 9 |
Hb_001456_060 |
0.0707839105 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 10 |
Hb_000339_040 |
0.071740519 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 11 |
Hb_002876_230 |
0.0718519102 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 12 |
Hb_001751_140 |
0.0730443071 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004236_050 |
0.0731265976 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 14 |
Hb_032264_060 |
0.0755933141 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 15 |
Hb_000331_110 |
0.0769220609 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 16 |
Hb_004052_080 |
0.0769995606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000390_230 |
0.0773950119 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 18 |
Hb_000144_070 |
0.0777717761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004837_180 |
0.0780851125 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_007007_120 |
0.0781737074 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |