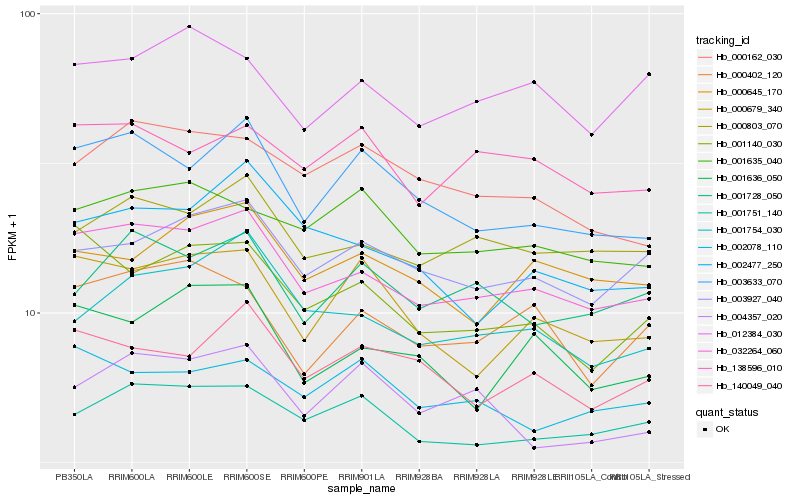
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032264_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 2 |
Hb_000803_070 |
0.0575948989 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 3 |
Hb_003633_070 |
0.0629549713 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 4 |
Hb_003927_040 |
0.0665936922 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 5 |
Hb_001140_030 |
0.0668190619 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 6 |
Hb_001751_140 |
0.0672549048 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001754_030 |
0.0673167296 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 8 |
Hb_001636_050 |
0.0689102093 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 9 |
Hb_140049_040 |
0.0710603682 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 10 |
Hb_000402_120 |
0.0724694681 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 11 |
Hb_002477_250 |
0.0733237614 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 12 |
Hb_000679_340 |
0.0735487767 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000645_170 |
0.0739529647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001728_050 |
0.075251521 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 15 |
Hb_012384_030 |
0.0755933141 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 16 |
Hb_001635_040 |
0.0760741624 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 17 |
Hb_000162_030 |
0.0767237023 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_138596_010 |
0.0771627132 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 19 |
Hb_004357_020 |
0.0777922695 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 20 |
Hb_002078_110 |
0.0783165829 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |