| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
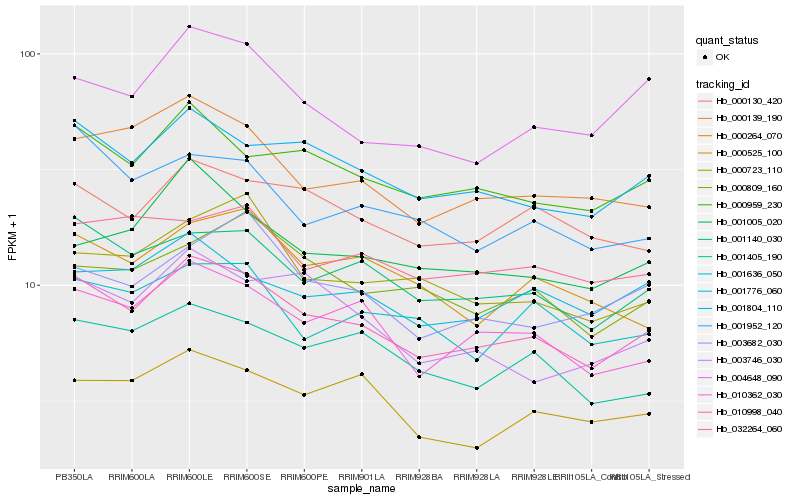
Hb_010998_040 |
0.0 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 2 |
Hb_001804_110 |
0.0605768739 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 3 |
Hb_001140_030 |
0.062716695 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 4 |
Hb_000959_230 |
0.0661524804 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 5 |
Hb_010362_030 |
0.067693901 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_000139_190 |
0.0763425878 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000130_420 |
0.0786981829 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 8 |
Hb_001405_190 |
0.0807109624 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_004648_090 |
0.0808509809 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 10 |
Hb_001952_120 |
0.0867564599 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001776_060 |
0.0868330732 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 12 |
Hb_000525_100 |
0.0879911874 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 13 |
Hb_001636_050 |
0.0884525949 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 14 |
Hb_001005_020 |
0.0886223392 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 15 |
Hb_032264_060 |
0.0894497999 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 16 |
Hb_003746_030 |
0.0894774652 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 17 |
Hb_000264_070 |
0.0894998783 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 18 |
Hb_000723_110 |
0.0902892904 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 19 |
Hb_003682_030 |
0.091398932 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 20 |
Hb_000809_160 |
0.0942406368 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |