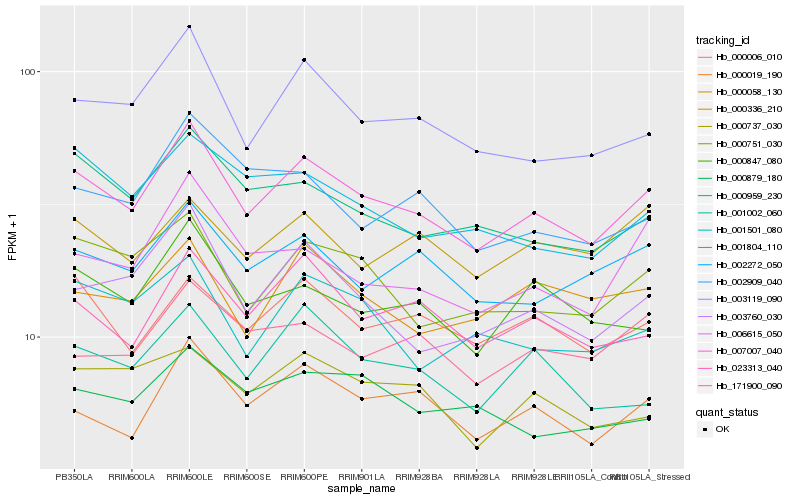
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007007_040 |
0.0 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 2 |
Hb_003119_090 |
0.0641521584 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 3 |
Hb_000019_190 |
0.065345409 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 4 |
Hb_000751_030 |
0.0686203416 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 5 |
Hb_023313_040 |
0.0693629218 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003760_030 |
0.0714037873 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 7 |
Hb_000006_010 |
0.073237255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002272_050 |
0.0733200025 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 9 |
Hb_000959_230 |
0.0740131677 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 10 |
Hb_000847_080 |
0.076612903 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 11 |
Hb_006615_050 |
0.0773612693 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 12 |
Hb_001002_060 |
0.0782676615 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 13 |
Hb_001501_080 |
0.0790972384 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000737_030 |
0.0808449372 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000058_130 |
0.0818207917 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 16 |
Hb_001804_110 |
0.0820927686 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 17 |
Hb_002909_040 |
0.0838122098 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 18 |
Hb_000879_180 |
0.083902782 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_171900_090 |
0.0843417049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000336_210 |
0.0845486586 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |