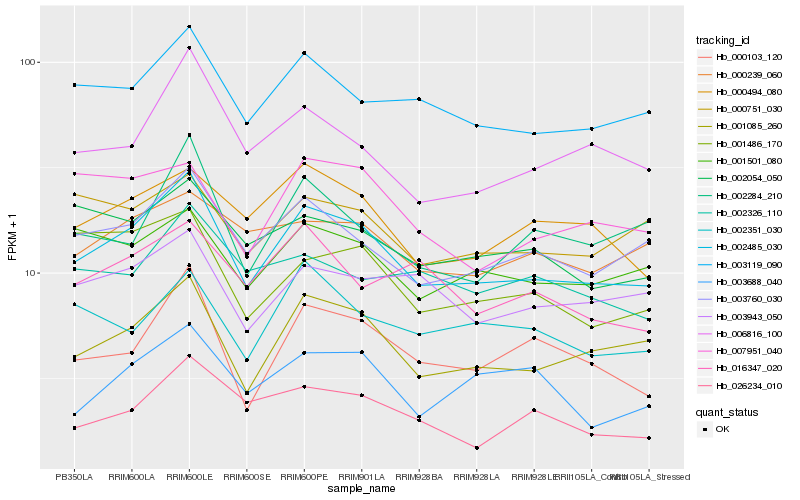
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002485_030 |
0.0 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 2 |
Hb_001085_260 |
0.0711467494 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 3 |
Hb_003760_030 |
0.081815255 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 4 |
Hb_003119_090 |
0.0878001016 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 5 |
Hb_006816_100 |
0.0949413376 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 6 |
Hb_000103_120 |
0.10053263 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000494_080 |
0.1012017004 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001501_080 |
0.1040247051 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003943_050 |
0.1062846838 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 10 |
Hb_002284_210 |
0.1080958717 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 11 |
Hb_002054_050 |
0.1101922819 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 12 |
Hb_026234_010 |
0.1102160364 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 13 |
Hb_000751_030 |
0.1117215911 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 14 |
Hb_000239_060 |
0.1119062556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001486_170 |
0.1132525699 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 16 |
Hb_007951_040 |
0.113490822 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 17 |
Hb_002351_030 |
0.1150676058 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_016347_020 |
0.1164808157 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 19 |
Hb_002326_110 |
0.1180627614 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 20 |
Hb_003688_040 |
0.1181740652 |
- |
- |
- |