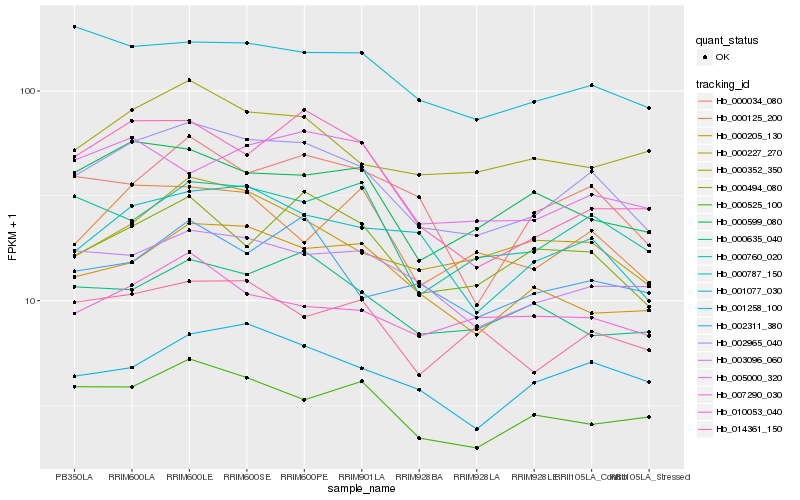
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002965_040 |
0.0 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 2 |
Hb_000787_150 |
0.0849386685 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 3 |
Hb_003096_060 |
0.0912751587 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 4 |
Hb_001258_100 |
0.0920039786 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000227_270 |
0.0927019771 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000494_080 |
0.094134226 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000205_130 |
0.0957227304 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000525_100 |
0.0967037679 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 9 |
Hb_000352_350 |
0.0968713455 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000034_080 |
0.0971686232 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000599_080 |
0.09772209 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 12 |
Hb_000760_020 |
0.0978139337 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_014361_150 |
0.0982231766 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 14 |
Hb_001077_030 |
0.1001888334 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000635_040 |
0.1009464465 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 16 |
Hb_000125_200 |
0.103361156 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 17 |
Hb_007290_030 |
0.1035218752 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 18 |
Hb_010053_040 |
0.1045735415 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 19 |
Hb_002311_380 |
0.1053320701 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 20 |
Hb_005000_320 |
0.1074189519 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |