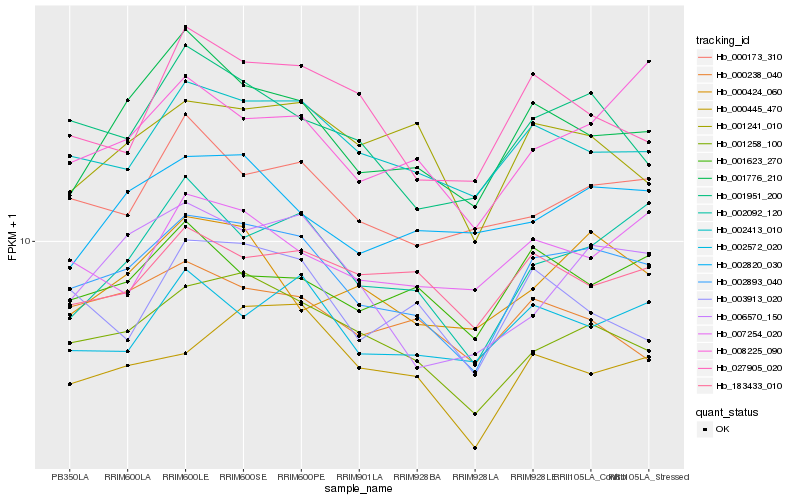
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002893_040 |
0.0 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001258_100 |
0.0719327108 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002413_010 |
0.0789499233 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_001951_200 |
0.0874883726 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001776_210 |
0.0913073041 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_002092_120 |
0.0969100309 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002572_020 |
0.0970271405 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 8 |
Hb_000173_310 |
0.0975690667 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000445_470 |
0.1004903847 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 10 |
Hb_001623_270 |
0.1005613362 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 11 |
Hb_183433_010 |
0.1016591768 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 12 |
Hb_006570_150 |
0.1023318767 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 13 |
Hb_027905_020 |
0.1036981881 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 14 |
Hb_001241_010 |
0.1038849263 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_007254_020 |
0.1057921152 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 16 |
Hb_008225_090 |
0.106940793 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000238_040 |
0.1070421722 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 18 |
Hb_000424_060 |
0.1071340234 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 19 |
Hb_002820_030 |
0.1076310544 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 20 |
Hb_003913_020 |
0.1078966034 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |