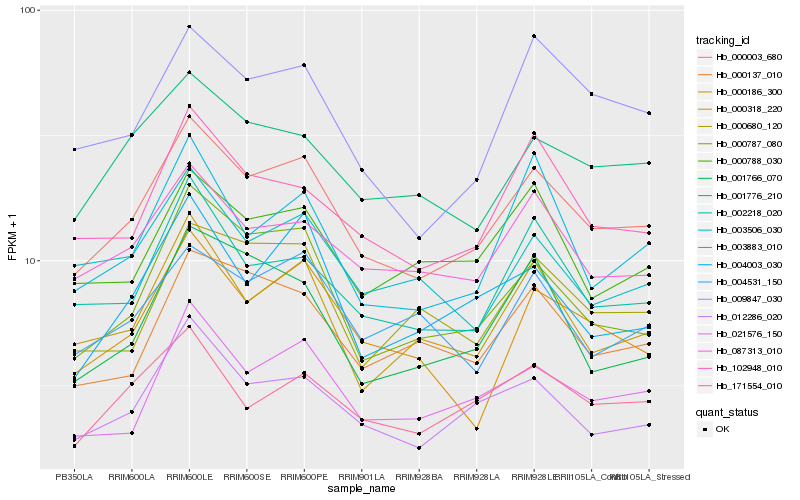
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_680 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 2 |
Hb_012286_020 |
0.0747093022 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 3 |
Hb_000787_080 |
0.0796383591 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 4 |
Hb_087313_010 |
0.0806138958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_021576_150 |
0.0812807178 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 6 |
Hb_002218_020 |
0.0820342371 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_102948_010 |
0.0827784186 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001776_210 |
0.0837748118 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000318_220 |
0.0842120878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_171554_010 |
0.089368563 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 11 |
Hb_009847_030 |
0.0903747066 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 12 |
Hb_000137_010 |
0.0910866942 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 13 |
Hb_000788_030 |
0.0933704499 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 14 |
Hb_004531_150 |
0.0956065094 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004003_030 |
0.0961295819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003883_010 |
0.0965084659 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 17 |
Hb_000680_120 |
0.097074459 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 18 |
Hb_003506_030 |
0.0985560462 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_000186_300 |
0.0987337726 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001766_070 |
0.0995963713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |