| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
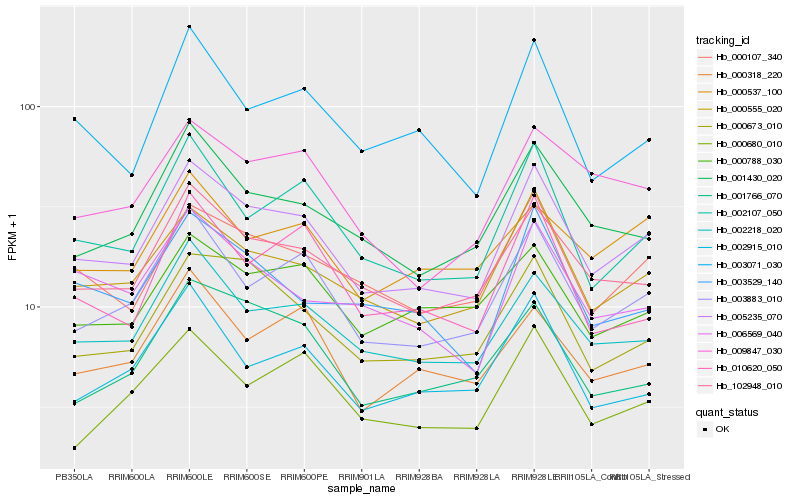
Hb_005235_070 |
0.0 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000555_020 |
0.067095357 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003883_010 |
0.0716384758 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 4 |
Hb_102948_010 |
0.0742386634 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000107_340 |
0.0754020479 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 6 |
Hb_002218_020 |
0.0770266319 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_002107_050 |
0.077268214 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 8 |
Hb_001430_020 |
0.0863692538 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000673_010 |
0.0875634556 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003071_030 |
0.09546627 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009847_030 |
0.0979028346 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 12 |
Hb_000788_030 |
0.1004431667 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_000318_220 |
0.1005714907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003529_140 |
0.1009395544 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002915_010 |
0.1032997999 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 16 |
Hb_006569_040 |
0.103551628 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 17 |
Hb_001766_070 |
0.1036536317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010620_050 |
0.1047228161 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 19 |
Hb_000680_010 |
0.105106644 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 20 |
Hb_000537_100 |
0.1054330409 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |