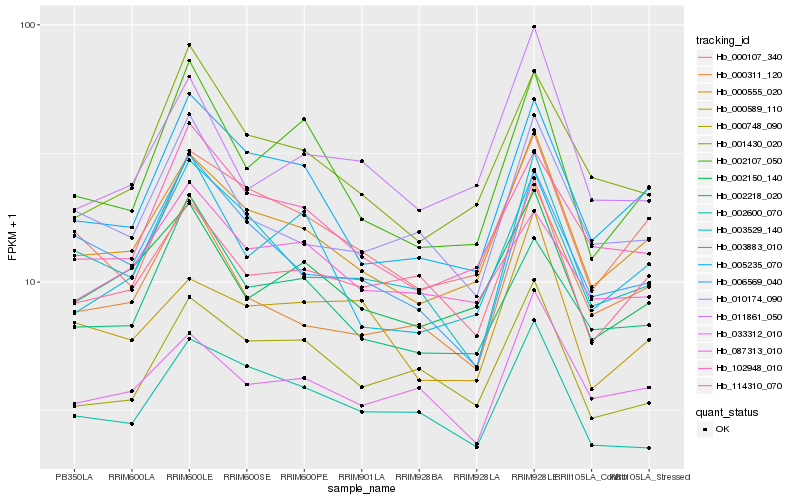
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000555_020 |
0.0 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000107_340 |
0.0595826143 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 3 |
Hb_005235_070 |
0.067095357 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002150_140 |
0.0768037056 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 5 |
Hb_102948_010 |
0.0805669436 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_114310_070 |
0.0848870157 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 7 |
Hb_000311_120 |
0.0879178859 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 8 |
Hb_001430_020 |
0.0882776524 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_003529_140 |
0.0914228987 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_033312_010 |
0.092133923 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_002107_050 |
0.092808264 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 12 |
Hb_002218_020 |
0.0948414748 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 13 |
Hb_003883_010 |
0.0951416501 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 14 |
Hb_002600_070 |
0.0954283188 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 15 |
Hb_000748_090 |
0.0962749739 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 16 |
Hb_010174_090 |
0.0963862391 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006569_040 |
0.0990604195 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 18 |
Hb_000589_110 |
0.0994765793 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 19 |
Hb_087313_010 |
0.1019235993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011861_050 |
0.1040130047 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |