| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
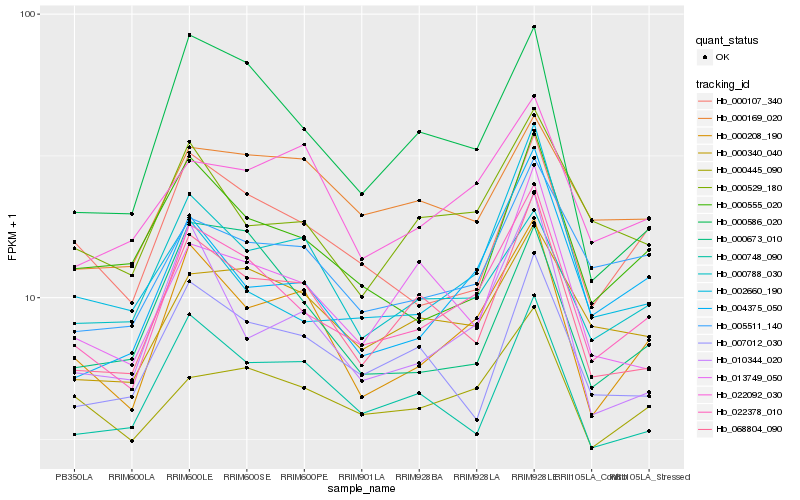
Hb_022378_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000208_190 |
0.0785037215 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 3 |
Hb_068804_090 |
0.0939401703 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 4 |
Hb_004375_050 |
0.0952559847 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000107_340 |
0.0969583103 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 6 |
Hb_000340_040 |
0.0973005923 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 7 |
Hb_005511_140 |
0.0985022771 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 8 |
Hb_000529_180 |
0.1006592206 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000748_090 |
0.1031668719 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 10 |
Hb_000445_090 |
0.1034964762 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 11 |
Hb_000555_020 |
0.10520298 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000586_020 |
0.1094110533 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 13 |
Hb_007012_030 |
0.1110751633 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_013749_050 |
0.1111164325 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010344_020 |
0.1121903544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 16 |
Hb_022092_030 |
0.1132181066 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000788_030 |
0.1136109203 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 18 |
Hb_000673_010 |
0.1143140326 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000169_020 |
0.1151846941 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 20 |
Hb_002660_190 |
0.1170894799 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |