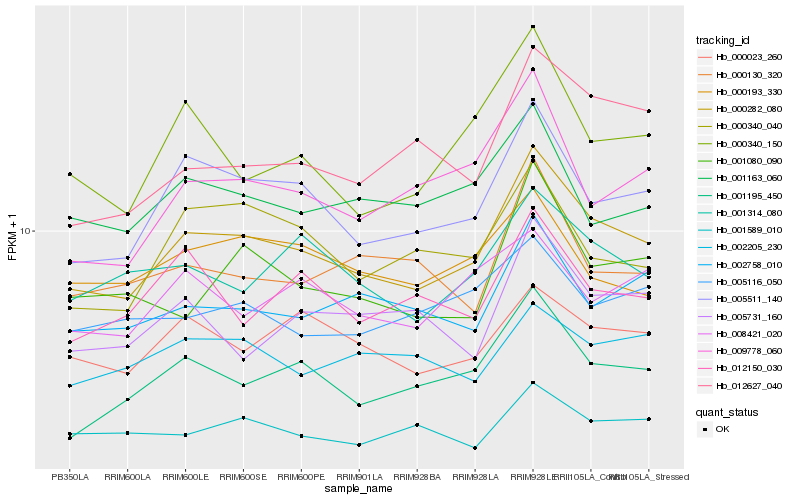
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000282_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005511_140 |
0.068196053 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 3 |
Hb_001195_450 |
0.0843206115 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 4 |
Hb_001314_080 |
0.0950331409 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 5 |
Hb_002758_010 |
0.0966151532 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 6 |
Hb_000023_260 |
0.0972523986 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001080_090 |
0.0997342522 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000340_150 |
0.0998257188 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 9 |
Hb_001163_060 |
0.1000633195 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 10 |
Hb_012627_040 |
0.1004430523 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 11 |
Hb_009778_060 |
0.1005332682 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 12 |
Hb_000340_040 |
0.1013006578 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 13 |
Hb_012150_030 |
0.1050948088 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000193_330 |
0.1054103994 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 15 |
Hb_001589_010 |
0.1061142608 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005116_050 |
0.1061606347 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_005731_160 |
0.1064337753 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_008421_020 |
0.1065457695 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000130_320 |
0.1070708344 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 20 |
Hb_002205_230 |
0.108302677 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |