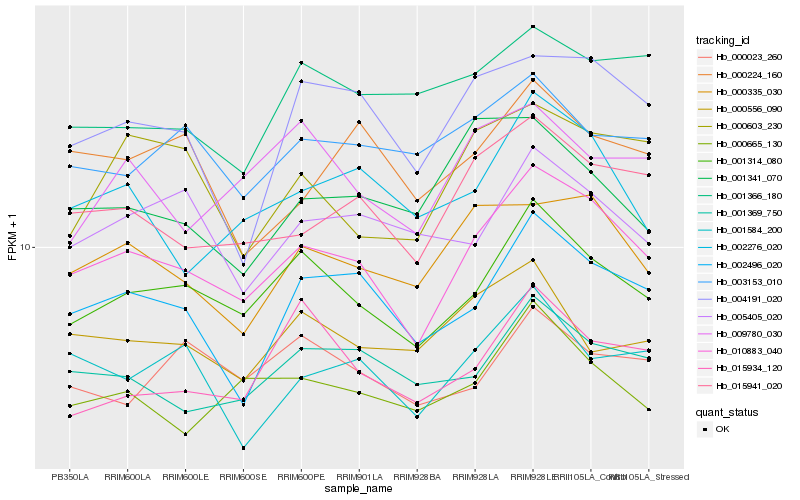
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_040 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 2 |
Hb_001314_080 |
0.068568748 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 3 |
Hb_015941_020 |
0.0797535036 |
- |
- |
- |
| 4 |
Hb_002496_020 |
0.089083621 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000335_030 |
0.0927098459 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 6 |
Hb_001369_750 |
0.0988494654 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 7 |
Hb_002276_020 |
0.1009924731 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004191_020 |
0.1026905374 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 9 |
Hb_000665_130 |
0.1038831647 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001584_200 |
0.1084699244 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 11 |
Hb_000603_230 |
0.1108989986 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 12 |
Hb_003153_010 |
0.1124363602 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 13 |
Hb_005405_020 |
0.1130874383 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_009780_030 |
0.1155044519 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 15 |
Hb_000224_160 |
0.1160217893 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_001341_070 |
0.1181363349 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000556_090 |
0.1183252092 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000023_260 |
0.1194189075 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001366_180 |
0.1196419907 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_015934_120 |
0.1202773295 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |