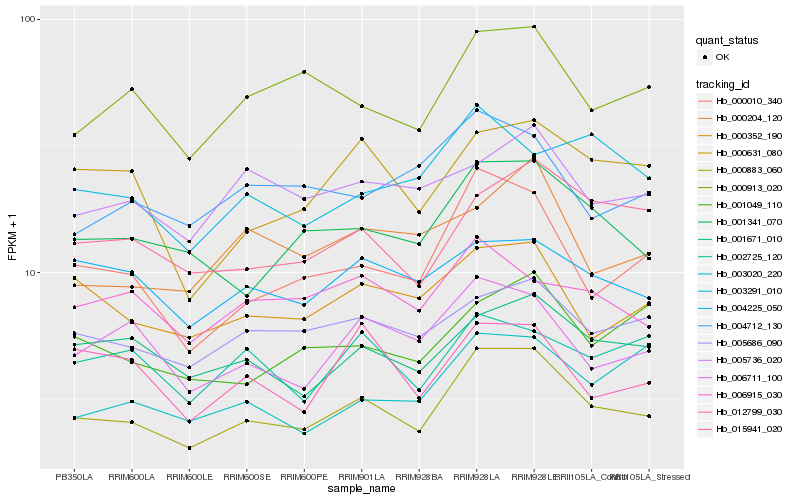
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000883_060 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 2 |
Hb_000010_340 |
0.0802604587 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001341_070 |
0.086718062 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001671_010 |
0.0930309878 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 5 |
Hb_015941_020 |
0.0940902337 |
- |
- |
- |
| 6 |
Hb_006711_100 |
0.0941119674 |
- |
- |
PREDICTED: uncharacterized protein LOC105637078 [Jatropha curcas] |
| 7 |
Hb_000913_020 |
0.0948205821 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000352_190 |
0.0953453334 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 9 |
Hb_005686_090 |
0.0965267493 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001049_110 |
0.1052971676 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000631_080 |
0.1056067718 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 12 |
Hb_004225_050 |
0.1070174409 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000204_120 |
0.109896965 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 14 |
Hb_005736_020 |
0.1102918983 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 15 |
Hb_004712_130 |
0.1108161295 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 16 |
Hb_003020_220 |
0.1110361626 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006915_030 |
0.1112064295 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |
| 18 |
Hb_012799_030 |
0.1116684519 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 19 |
Hb_002725_120 |
0.1121736361 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003291_010 |
0.1141192802 |
- |
- |
ATP-dependent protease La, putative [Ricinus communis] |