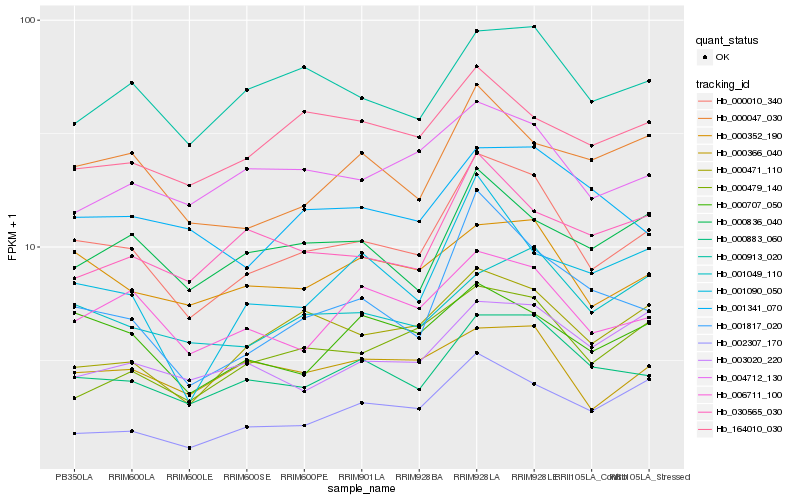
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_340 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000883_060 |
0.0802604587 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 3 |
Hb_006711_100 |
0.1033204109 |
- |
- |
PREDICTED: uncharacterized protein LOC105637078 [Jatropha curcas] |
| 4 |
Hb_000352_190 |
0.1040804307 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 5 |
Hb_001817_020 |
0.1052525625 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 6 |
Hb_000913_020 |
0.1066697368 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000471_110 |
0.1068821455 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000047_030 |
0.1111193608 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 9 |
Hb_000836_040 |
0.1119845935 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000707_050 |
0.1123543856 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004712_130 |
0.1130109922 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 12 |
Hb_001049_110 |
0.1140158039 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000479_140 |
0.1140521615 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 14 |
Hb_000366_040 |
0.1144161424 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 15 |
Hb_001090_050 |
0.1191374693 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_030565_030 |
0.1193034155 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 17 |
Hb_001341_070 |
0.1199231447 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002307_170 |
0.1202800526 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 19 |
Hb_164010_030 |
0.1220013507 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 20 |
Hb_003020_220 |
0.1249356123 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |