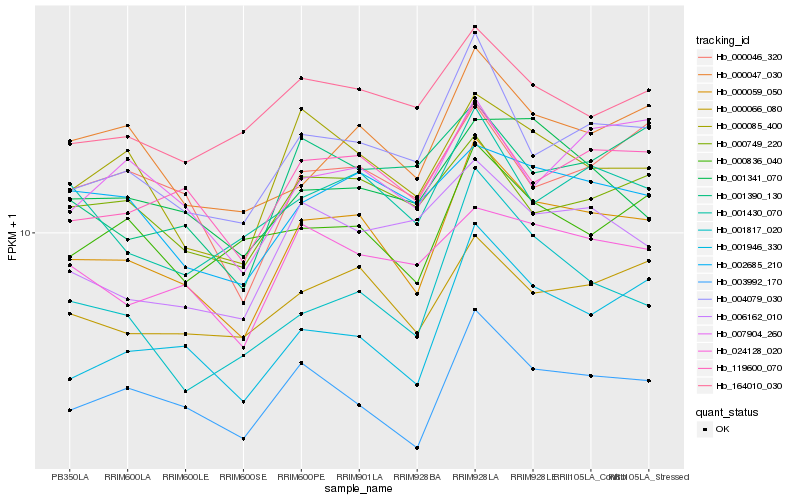
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 2 |
Hb_004079_030 |
0.0870502552 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003992_170 |
0.0889552718 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 4 |
Hb_119600_070 |
0.093076949 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 5 |
Hb_001946_330 |
0.0947824673 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 6 |
Hb_000066_080 |
0.0992973034 |
- |
- |
PREDICTED: mRNA cap guanine-N7 methyltransferase 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000047_030 |
0.1031107637 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 8 |
Hb_000085_400 |
0.105291264 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 9 |
Hb_024128_020 |
0.1089072744 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001430_070 |
0.1096558246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002685_210 |
0.113544811 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007904_260 |
0.1150898984 |
- |
- |
PREDICTED: uncharacterized protein LOC105644715 [Jatropha curcas] |
| 13 |
Hb_000046_320 |
0.1152190927 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_006162_010 |
0.1157610381 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |
| 15 |
Hb_000749_220 |
0.1163117514 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000836_040 |
0.1164921899 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_164010_030 |
0.1177186817 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 18 |
Hb_001341_070 |
0.118425679 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001390_130 |
0.1195581729 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 20 |
Hb_001817_020 |
0.1195695453 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |