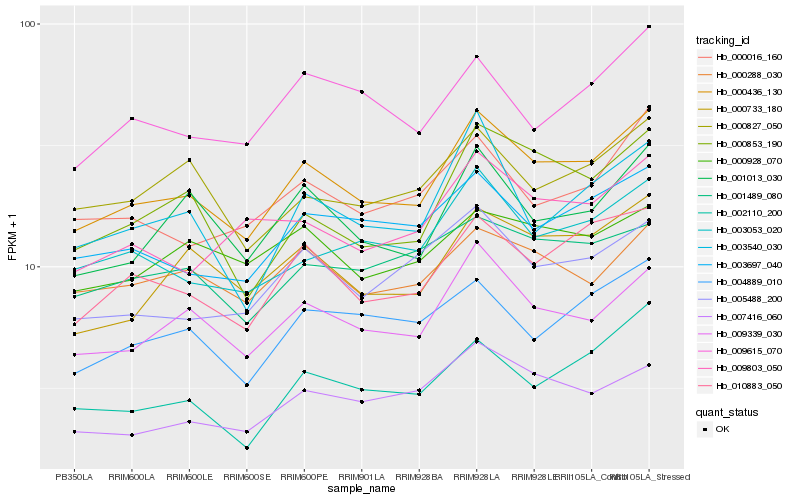
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000436_130 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 2 |
Hb_009339_030 |
0.0592775997 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 3 |
Hb_010883_050 |
0.0616293428 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000733_180 |
0.0716006391 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 5 |
Hb_003697_040 |
0.0725565525 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 6 |
Hb_001013_030 |
0.0725645469 |
- |
- |
GTP-binding protein yptv3, putative [Ricinus communis] |
| 7 |
Hb_003053_020 |
0.0734551046 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 8 |
Hb_005488_200 |
0.0780655854 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004889_010 |
0.0798117756 |
- |
- |
Vacuolar protein sorting-associated protein-like protein [Medicago truncatula] |
| 10 |
Hb_000288_030 |
0.0798367072 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 11 |
Hb_007416_060 |
0.0803447612 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 18 [Jatropha curcas] |
| 12 |
Hb_000016_160 |
0.0811293698 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 13 |
Hb_000827_050 |
0.0815893631 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 14 |
Hb_000853_190 |
0.0826709772 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 15 |
Hb_001489_080 |
0.0830805998 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_009615_070 |
0.084056672 |
- |
- |
type X protein phosphatase-II [Populus trichocarpa] |
| 17 |
Hb_009803_050 |
0.0841270616 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 18 |
Hb_003540_030 |
0.0846389939 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000928_070 |
0.0847778407 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002110_200 |
0.0848919973 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |