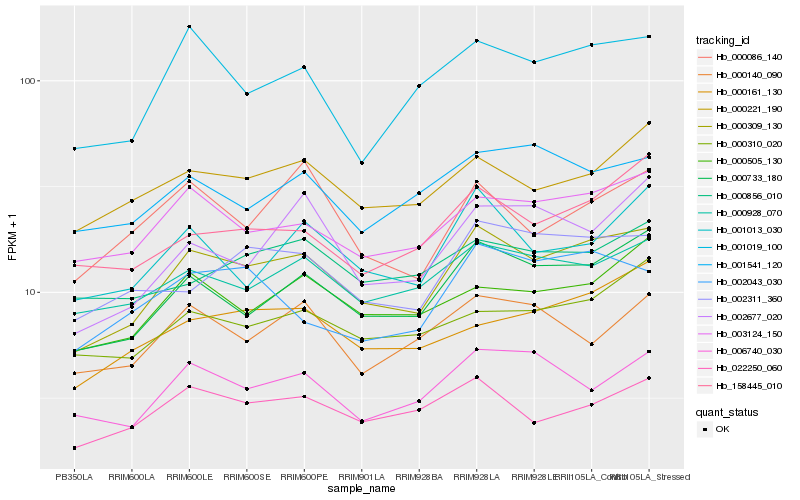
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000309_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_09592 [Jatropha curcas] |
| 2 |
Hb_000733_180 |
0.0665324987 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 3 |
Hb_003124_150 |
0.0841306136 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 4 |
Hb_000928_070 |
0.0906643089 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_022250_060 |
0.0919895819 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001019_100 |
0.0926247401 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 7 |
Hb_000221_190 |
0.0940053614 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 8 |
Hb_002311_360 |
0.0950606344 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 9 |
Hb_000505_130 |
0.0951616254 |
- |
- |
PREDICTED: uncharacterized protein LOC105641262 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001013_030 |
0.0975789056 |
- |
- |
GTP-binding protein yptv3, putative [Ricinus communis] |
| 11 |
Hb_006740_030 |
0.0976355447 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_158445_010 |
0.1010703344 |
- |
- |
PREDICTED: cysteine protease ATG4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000161_130 |
0.1013651441 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000310_020 |
0.1025472522 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 15 |
Hb_002677_020 |
0.104201062 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 16 |
Hb_001541_120 |
0.1049133502 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 17 |
Hb_000086_140 |
0.105721428 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 18 |
Hb_002043_030 |
0.1059540113 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 19 |
Hb_000140_090 |
0.1060662039 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 20 |
Hb_000856_010 |
0.1077471631 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |