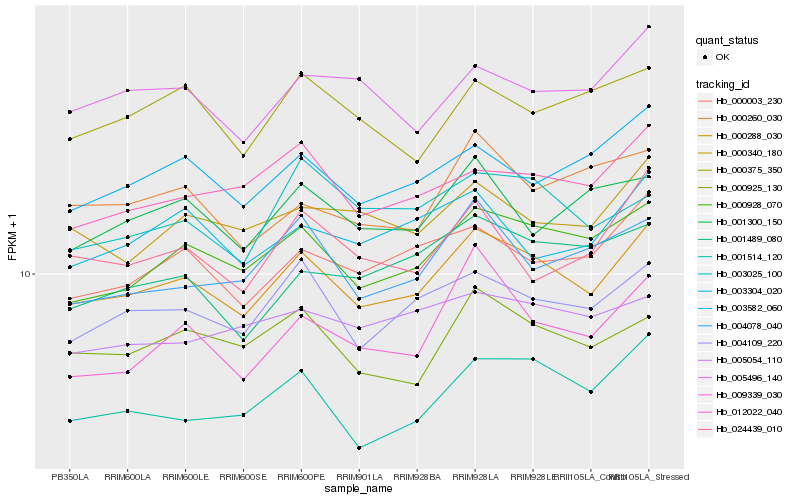
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000288_030 |
0.0 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 2 |
Hb_000928_070 |
0.0517894617 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_004109_220 |
0.0575326703 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 4 |
Hb_000003_230 |
0.0653830256 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000925_130 |
0.0656922124 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_003582_060 |
0.0687430983 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_012022_040 |
0.0700968095 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 8 |
Hb_001514_120 |
0.0705443403 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 9 |
Hb_000340_180 |
0.0713344455 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of NFKB 1 [Jatropha curcas] |
| 10 |
Hb_004078_040 |
0.0732595302 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 11 |
Hb_005496_140 |
0.0737066803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009339_030 |
0.0760606524 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 13 |
Hb_001300_150 |
0.0766574693 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 14 |
Hb_003304_020 |
0.0772359573 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 15 |
Hb_000260_030 |
0.0773533195 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 16 |
Hb_003025_100 |
0.0777281921 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_024439_010 |
0.0777317 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 18 |
Hb_000375_350 |
0.077832442 |
- |
- |
PREDICTED: uncharacterized protein LOC105641632 [Jatropha curcas] |
| 19 |
Hb_001489_080 |
0.0780544189 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_005054_110 |
0.0795950396 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |