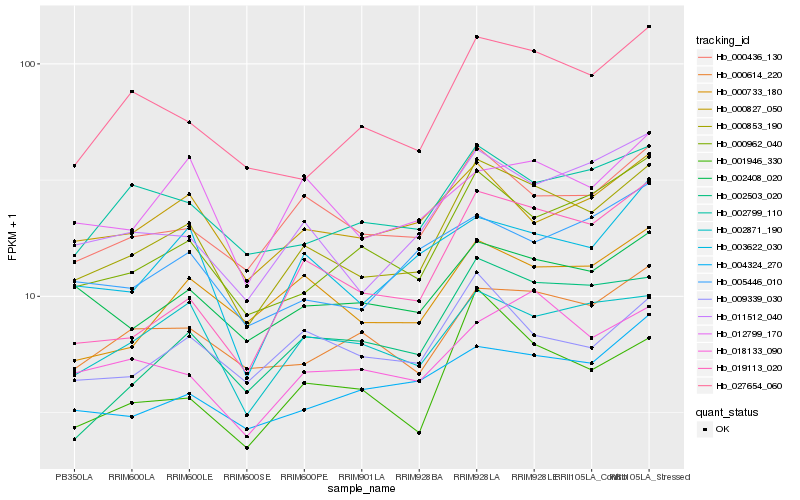
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_190 |
0.0 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 2 |
Hb_000436_130 |
0.0826709772 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 3 |
Hb_011512_040 |
0.0855133331 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 4 |
Hb_000962_040 |
0.0900936427 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000614_220 |
0.0948485873 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 6 |
Hb_027654_060 |
0.0953853901 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 7 |
Hb_002871_190 |
0.0957138672 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |
| 8 |
Hb_001946_330 |
0.096281104 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 9 |
Hb_002408_020 |
0.0974466534 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 10 |
Hb_002799_110 |
0.0984453694 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 11 |
Hb_000733_180 |
0.0987711343 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 12 |
Hb_019113_020 |
0.0994102098 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 13 |
Hb_003622_030 |
0.099486221 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_009339_030 |
0.1011986132 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 15 |
Hb_018133_090 |
0.1012621941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004324_270 |
0.1032762789 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |
| 17 |
Hb_000827_050 |
0.1046034111 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 18 |
Hb_002503_020 |
0.1061898161 |
- |
- |
Phosphatase 2C family protein, putative [Theobroma cacao] |
| 19 |
Hb_012799_170 |
0.1074722264 |
- |
- |
PREDICTED: protein GLC8 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005446_010 |
0.1081190234 |
- |
- |
PREDICTED: zinc finger protein 830 [Jatropha curcas] |