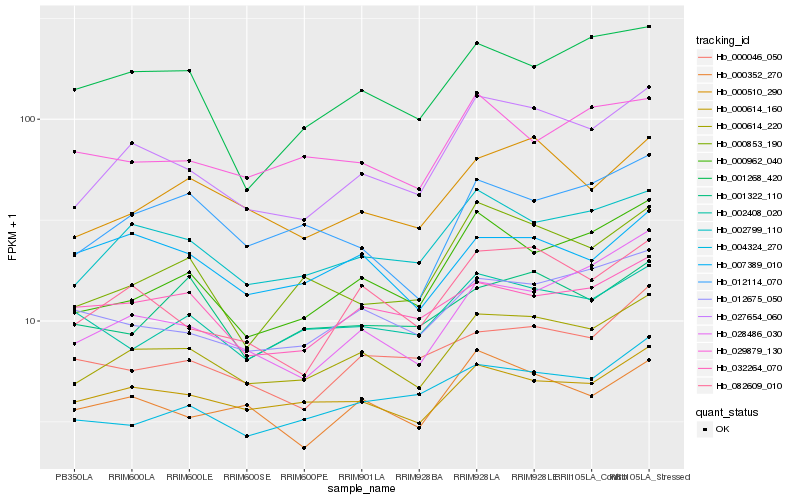
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000614_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 2 |
Hb_002799_110 |
0.0607448362 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 3 |
Hb_027654_060 |
0.0665083935 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 4 |
Hb_000614_160 |
0.0681735638 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04130, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000510_290 |
0.0728378012 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_012675_050 |
0.0808640598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012114_070 |
0.083531529 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 8 |
Hb_082609_010 |
0.0843160219 |
- |
- |
PREDICTED: uncharacterized protein LOC105644299 [Jatropha curcas] |
| 9 |
Hb_000962_040 |
0.0844050493 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_032264_070 |
0.0870463875 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004324_270 |
0.0879179184 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |
| 12 |
Hb_002408_020 |
0.0895886659 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 13 |
Hb_000046_050 |
0.0905949008 |
- |
- |
PREDICTED: LEC14B homolog [Jatropha curcas] |
| 14 |
Hb_000352_270 |
0.0916036722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001322_110 |
0.0938490701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028486_030 |
0.0944957556 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 17 |
Hb_007389_010 |
0.0945838834 |
- |
- |
PREDICTED: THO complex subunit 7A [Jatropha curcas] |
| 18 |
Hb_000853_190 |
0.0948485873 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 19 |
Hb_001268_420 |
0.0956229534 |
- |
- |
PREDICTED: uncharacterized protein LOC105630158 [Jatropha curcas] |
| 20 |
Hb_029879_130 |
0.0959138248 |
- |
- |
Derlin-2, putative [Ricinus communis] |