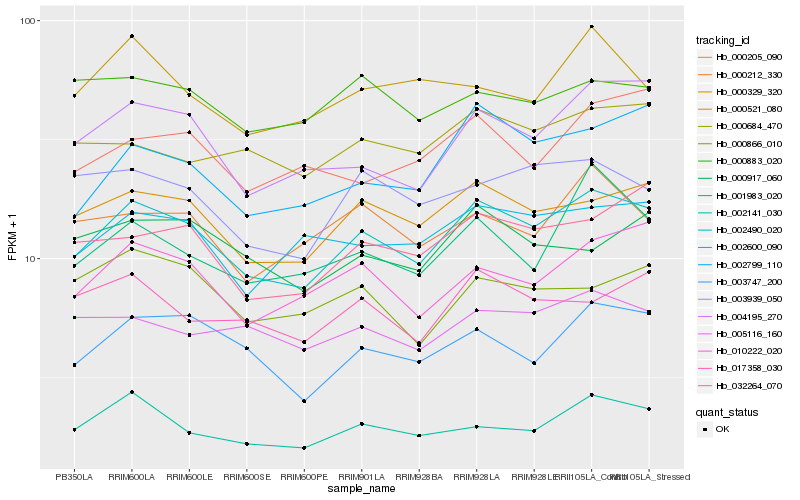
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002490_020 |
0.0 |
- |
- |
peptidyl-tRNA hydrolase family protein [Populus trichocarpa] |
| 2 |
Hb_000329_320 |
0.0627840563 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 3 |
Hb_004195_270 |
0.0750849224 |
- |
- |
Ubiquitin-conjugating enzyme 16 [Theobroma cacao] |
| 4 |
Hb_003747_200 |
0.0765625061 |
- |
- |
PREDICTED: DNA cross-link repair protein SNM1 [Jatropha curcas] |
| 5 |
Hb_010222_020 |
0.0775363092 |
- |
- |
PREDICTED: uncharacterized protein LOC105647642 [Jatropha curcas] |
| 6 |
Hb_002141_030 |
0.0776229244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002600_090 |
0.080588845 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002799_110 |
0.0837194791 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 9 |
Hb_032264_070 |
0.0846125811 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000212_330 |
0.0883877032 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_005116_160 |
0.0886327394 |
transcription factor |
TF Family: TRAF |
- |
| 12 |
Hb_003939_050 |
0.0886802031 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000684_470 |
0.0895428593 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 14 |
Hb_001983_020 |
0.0897440052 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 15 |
Hb_017358_030 |
0.0900931337 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000917_060 |
0.0943597111 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 17 |
Hb_000205_090 |
0.0955890059 |
- |
- |
hypothetical protein POPTR_0019s03570g [Populus trichocarpa] |
| 18 |
Hb_000883_020 |
0.0960817171 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 19 |
Hb_000866_010 |
0.0962928552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000521_080 |
0.0963821737 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |