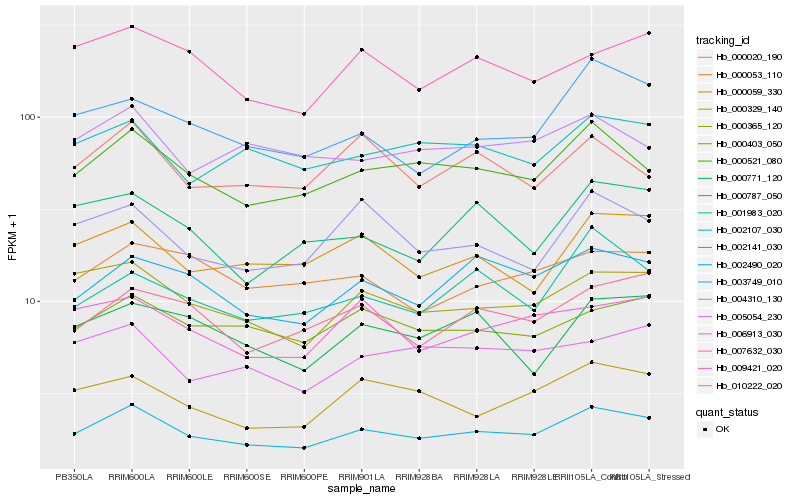
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002141_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000521_080 |
0.0619032807 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |
| 3 |
Hb_000365_120 |
0.0689937798 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_002490_020 |
0.0776229244 |
- |
- |
peptidyl-tRNA hydrolase family protein [Populus trichocarpa] |
| 5 |
Hb_000403_050 |
0.0813613662 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002107_030 |
0.0831927605 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000059_330 |
0.0839236724 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_010222_020 |
0.0850324293 |
- |
- |
PREDICTED: uncharacterized protein LOC105647642 [Jatropha curcas] |
| 9 |
Hb_004310_130 |
0.0876774532 |
- |
- |
PREDICTED: uncharacterized protein LOC105643225 [Jatropha curcas] |
| 10 |
Hb_007632_030 |
0.0879837458 |
- |
- |
PREDICTED: poly(A)-specific ribonuclease PARN isoform X1 [Jatropha curcas] |
| 11 |
Hb_000329_140 |
0.0883233498 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 12 |
Hb_006913_030 |
0.0893723566 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 13 |
Hb_009421_020 |
0.0898762692 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 14 |
Hb_000053_110 |
0.091794643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001983_020 |
0.0925407082 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 16 |
Hb_005054_230 |
0.0926936788 |
- |
- |
PREDICTED: wiskott-Aldrich syndrome protein family member 2 [Jatropha curcas] |
| 17 |
Hb_003749_010 |
0.0930487752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000020_190 |
0.093180522 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 19 |
Hb_000787_050 |
0.0951896623 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000771_120 |
0.0952739852 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |