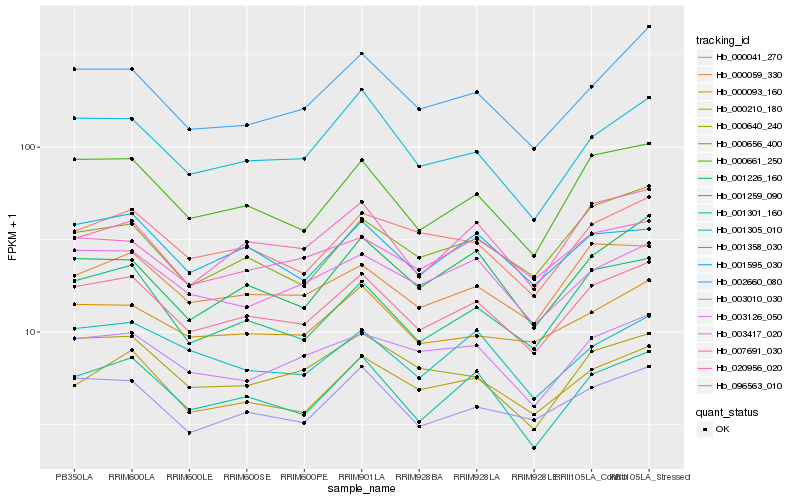
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096563_010 |
0.0 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 2 |
Hb_000661_250 |
0.050379252 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 3 |
Hb_003010_030 |
0.0526388566 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 4 |
Hb_001301_160 |
0.0545654041 |
- |
- |
PREDICTED: CRS2-associated factor 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001259_090 |
0.0547568661 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 6 |
Hb_000059_330 |
0.0563430112 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_000210_180 |
0.0582518428 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_000093_160 |
0.059238169 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001358_030 |
0.060836126 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 10 |
Hb_000041_270 |
0.0613438342 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 11 |
Hb_020956_020 |
0.0630069539 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 12 |
Hb_002660_080 |
0.0635886168 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 13 |
Hb_001595_030 |
0.0639876453 |
- |
- |
PREDICTED: nuclear inhibitor of protein phosphatase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003417_020 |
0.0645626226 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003126_050 |
0.064598997 |
- |
- |
PREDICTED: cell cycle checkpoint protein RAD17 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001226_160 |
0.0653851322 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000640_240 |
0.0664794301 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 18 |
Hb_001305_010 |
0.0669696679 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000656_400 |
0.0673986878 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 20 |
Hb_007691_030 |
0.0679913723 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |