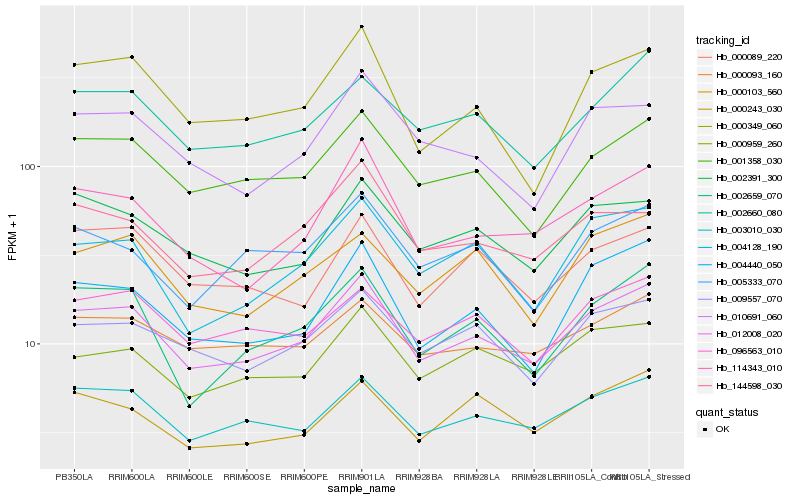
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012008_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104233229 [Nicotiana sylvestris] |
| 2 |
Hb_003010_030 |
0.0591865027 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 3 |
Hb_001358_030 |
0.0600441422 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 4 |
Hb_009557_070 |
0.0689956621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000959_260 |
0.0709375922 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 6 |
Hb_002660_080 |
0.071583925 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 7 |
Hb_002391_300 |
0.0734181324 |
- |
- |
hypothetical protein PHAVU_002G104100g [Phaseolus vulgaris] |
| 8 |
Hb_002659_070 |
0.0745823306 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 9 |
Hb_096563_010 |
0.0748059289 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 10 |
Hb_114343_010 |
0.0760057709 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 11 |
Hb_004128_190 |
0.0761196836 |
- |
- |
PREDICTED: dirigent protein 17-like [Jatropha curcas] |
| 12 |
Hb_000093_160 |
0.0763415165 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004440_050 |
0.077926262 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 14 |
Hb_000349_060 |
0.080101532 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 15 |
Hb_000103_560 |
0.0808734988 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 16 |
Hb_000089_220 |
0.0812784995 |
- |
- |
PREDICTED: protein FAM133-like [Populus euphratica] |
| 17 |
Hb_144598_030 |
0.0814012273 |
- |
- |
PREDICTED: mitotic apparatus protein p62 [Jatropha curcas] |
| 18 |
Hb_005333_070 |
0.0814388196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010691_060 |
0.0819244718 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 20 |
Hb_000243_030 |
0.0820912654 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |