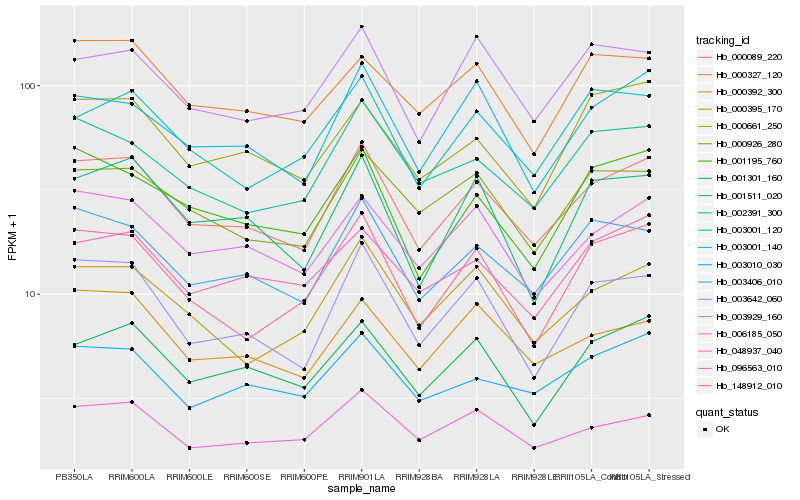
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000089_220 |
0.0 |
- |
- |
PREDICTED: protein FAM133-like [Populus euphratica] |
| 2 |
Hb_003406_010 |
0.0542490962 |
- |
- |
putative ATP-dependent RNA helicase DHX36 [Morus notabilis] |
| 3 |
Hb_006185_050 |
0.0588809879 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 4 |
Hb_003642_060 |
0.0609780634 |
- |
- |
hypothetical protein RCOM_1189860 [Ricinus communis] |
| 5 |
Hb_003001_140 |
0.065155268 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 6 |
Hb_003010_030 |
0.0654277521 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 7 |
Hb_000926_280 |
0.0668565981 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 8 |
Hb_048937_040 |
0.0682509777 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_002391_300 |
0.0685362188 |
- |
- |
hypothetical protein PHAVU_002G104100g [Phaseolus vulgaris] |
| 10 |
Hb_001301_160 |
0.0687262634 |
- |
- |
PREDICTED: CRS2-associated factor 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000395_170 |
0.0687912662 |
- |
- |
PREDICTED: probable transcriptional regulatory protein At2g25830 [Jatropha curcas] |
| 12 |
Hb_000392_300 |
0.0693488889 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 13 |
Hb_001195_760 |
0.0696685908 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_148912_010 |
0.0705621288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_096563_010 |
0.0714997758 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 16 |
Hb_000327_120 |
0.0734322241 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 17 |
Hb_003001_120 |
0.0746396219 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000661_250 |
0.0746986171 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 19 |
Hb_003929_160 |
0.0749666832 |
- |
- |
PREDICTED: pyruvate dehydrogenase (acetyl-transferring) kinase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001511_020 |
0.0750757428 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |