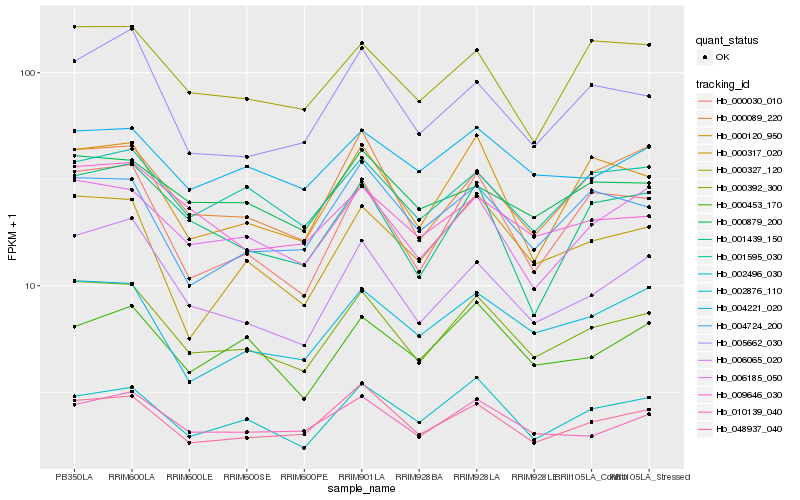
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_300 |
0.0 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 2 |
Hb_006065_020 |
0.0582313913 |
- |
- |
- |
| 3 |
Hb_006185_050 |
0.0587105527 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 4 |
Hb_048937_040 |
0.0623855349 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 5 |
Hb_000030_010 |
0.0684164843 |
- |
- |
PREDICTED: uncharacterized protein LOC105632627 [Jatropha curcas] |
| 6 |
Hb_000089_220 |
0.0693488889 |
- |
- |
PREDICTED: protein FAM133-like [Populus euphratica] |
| 7 |
Hb_002496_030 |
0.0697704456 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 8 |
Hb_010139_040 |
0.0722069817 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002876_110 |
0.0722394386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001595_030 |
0.0722582938 |
- |
- |
PREDICTED: nuclear inhibitor of protein phosphatase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000327_120 |
0.0726746855 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 12 |
Hb_009646_030 |
0.075132356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000120_950 |
0.0762610116 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 14 |
Hb_000879_200 |
0.0765082937 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 15 |
Hb_005662_030 |
0.0789203869 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_004221_020 |
0.0791594721 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_004724_200 |
0.0805703174 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 18 |
Hb_001439_150 |
0.0819736898 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000317_020 |
0.0822454039 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 20 |
Hb_000453_170 |
0.0832779225 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |