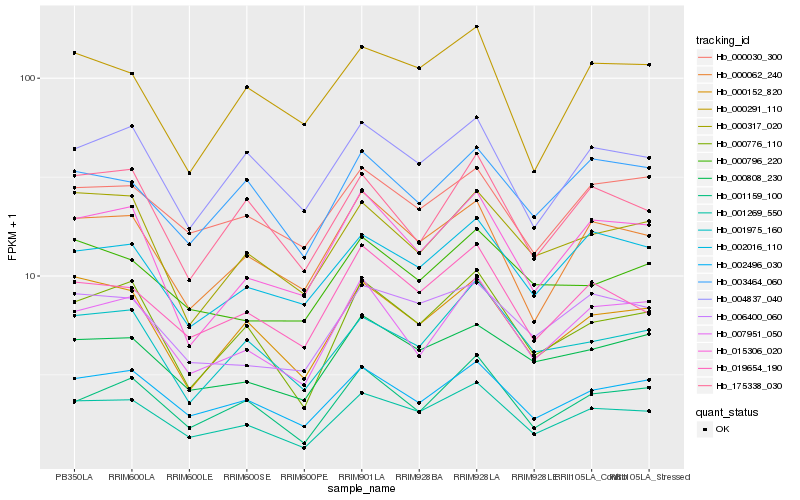
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_550 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s03510g [Populus trichocarpa] |
| 2 |
Hb_002496_030 |
0.0543193295 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 3 |
Hb_019654_190 |
0.0656826149 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000776_110 |
0.0671048115 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000152_820 |
0.0696291334 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial [Jatropha curcas] |
| 6 |
Hb_006400_060 |
0.0698166776 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002016_110 |
0.0706914493 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 8 |
Hb_000062_240 |
0.0716592584 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 9 |
Hb_000808_230 |
0.0727179856 |
- |
- |
PREDICTED: peptide chain release factor PrfB2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004837_040 |
0.0729743096 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 11 |
Hb_000030_300 |
0.0765492699 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 12 |
Hb_001975_160 |
0.0772998609 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001159_100 |
0.0785689032 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g22760 [Jatropha curcas] |
| 14 |
Hb_007951_050 |
0.0791539402 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003464_060 |
0.081789921 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 16 |
Hb_000317_020 |
0.0825018115 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 17 |
Hb_015306_020 |
0.0830109671 |
transcription factor |
TF Family: MYB-related |
PREDICTED: dnaJ homolog subfamily C member 2 [Jatropha curcas] |
| 18 |
Hb_000796_220 |
0.0843305234 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 19 |
Hb_175338_030 |
0.0849783317 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 20 |
Hb_000291_110 |
0.086026462 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |