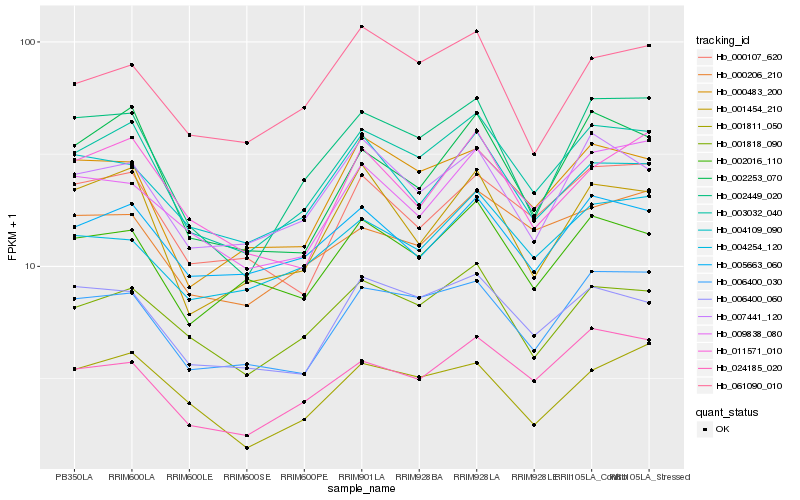
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003032_040 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 2 |
Hb_001818_090 |
0.0614031284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006400_030 |
0.0638082225 |
- |
- |
hypothetical protein OsI_01237 [Oryza sativa Indica Group] |
| 4 |
Hb_006400_060 |
0.0652053888 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000483_200 |
0.0662086808 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_024185_020 |
0.0688306106 |
- |
- |
- |
| 7 |
Hb_002449_020 |
0.0692750834 |
- |
- |
PREDICTED: tRNA (guanine(9)-N1)-methyltransferase [Cucumis sativus] |
| 8 |
Hb_007441_120 |
0.0697221001 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 9 |
Hb_002016_110 |
0.072320707 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 10 |
Hb_005663_060 |
0.0752355335 |
- |
- |
PREDICTED: uncharacterized protein LOC105770600 [Gossypium raimondii] |
| 11 |
Hb_002253_070 |
0.0755551327 |
transcription factor |
TF Family: NAC |
NAC transcription factor 042 [Jatropha curcas] |
| 12 |
Hb_000206_210 |
0.0782706602 |
- |
- |
PREDICTED: G patch domain-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_000107_620 |
0.0797070158 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 14 |
Hb_001454_210 |
0.0798154721 |
- |
- |
PREDICTED: uncharacterized protein LOC105643435 [Jatropha curcas] |
| 15 |
Hb_004109_090 |
0.0798899588 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_009838_080 |
0.0809985946 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 17 |
Hb_004254_120 |
0.0816422637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_061090_010 |
0.0836391859 |
- |
- |
RAN BINDING protein 1 [Populus trichocarpa] |
| 19 |
Hb_011571_010 |
0.0845034288 |
- |
- |
Metal tolerance protein C4, putative [Ricinus communis] |
| 20 |
Hb_001811_050 |
0.0848775813 |
- |
- |
PREDICTED: protein EXECUTER 1, chloroplastic [Jatropha curcas] |