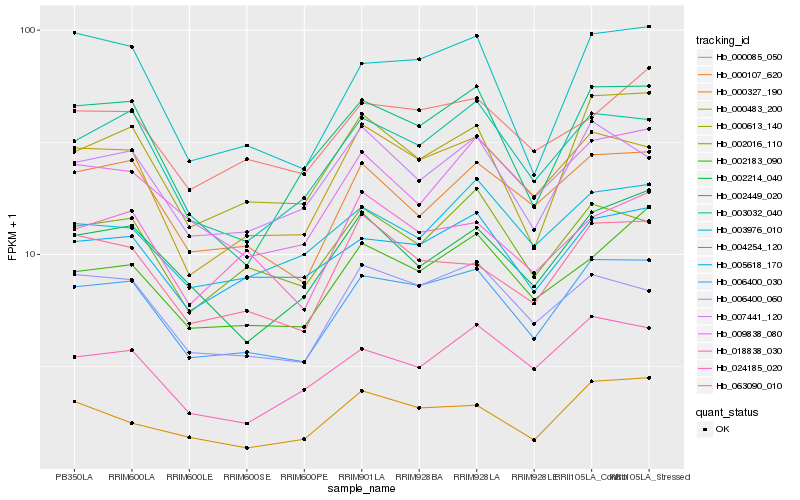
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006400_030 |
0.0 |
- |
- |
hypothetical protein OsI_01237 [Oryza sativa Indica Group] |
| 2 |
Hb_000483_200 |
0.0622816843 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003032_040 |
0.0638082225 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 4 |
Hb_006400_060 |
0.0689150732 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005618_170 |
0.0699192893 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 6 |
Hb_003976_010 |
0.0708191255 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 7 |
Hb_000107_620 |
0.0715507369 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 8 |
Hb_063090_010 |
0.0735519664 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002016_110 |
0.0754041618 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 10 |
Hb_000613_140 |
0.0764381086 |
- |
- |
PREDICTED: indole-3-glycerol phosphate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009838_080 |
0.0766380579 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 12 |
Hb_002183_090 |
0.0774852469 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 13 |
Hb_007441_120 |
0.0776601017 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000085_050 |
0.0789032197 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_024185_020 |
0.0811692461 |
- |
- |
- |
| 16 |
Hb_002449_020 |
0.081489121 |
- |
- |
PREDICTED: tRNA (guanine(9)-N1)-methyltransferase [Cucumis sativus] |
| 17 |
Hb_018838_030 |
0.0818558722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002214_040 |
0.0821920886 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_004254_120 |
0.0828804822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000327_190 |
0.0829741246 |
- |
- |
PREDICTED: CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase 2-like [Jatropha curcas] |