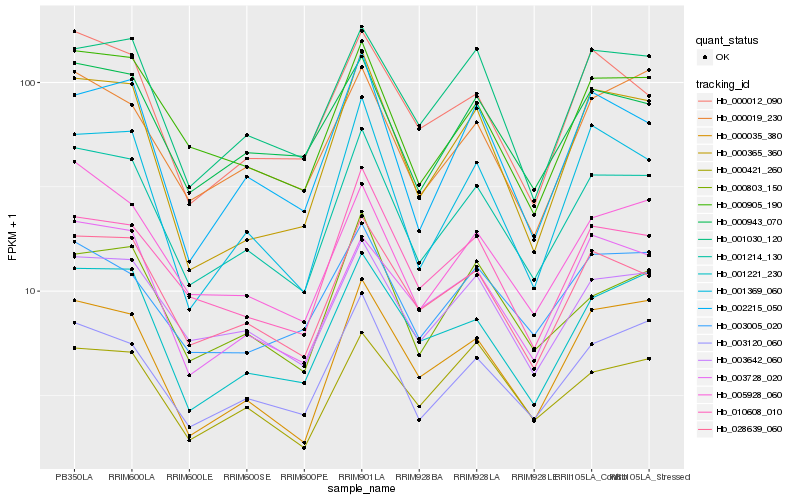
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001214_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646578 [Jatropha curcas] |
| 2 |
Hb_028639_060 |
0.0548917249 |
- |
- |
transporter, putative [Ricinus communis] |
| 3 |
Hb_003642_060 |
0.0561299954 |
- |
- |
hypothetical protein RCOM_1189860 [Ricinus communis] |
| 4 |
Hb_000905_190 |
0.0610932388 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 5 |
Hb_003120_060 |
0.0640078731 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 6 |
Hb_000943_070 |
0.0676028963 |
- |
- |
PREDICTED: uncharacterized protein LOC105635592 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000803_150 |
0.067891491 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001030_120 |
0.0717070935 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_005928_060 |
0.0737675903 |
- |
- |
DNA-directed RNA polymerase III 25 kD polypeptide, putative [Ricinus communis] |
| 10 |
Hb_010608_010 |
0.0750118423 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 11 |
Hb_001221_230 |
0.0765456258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002215_050 |
0.0772877188 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 13 |
Hb_001369_060 |
0.0784433544 |
- |
- |
PREDICTED: uncharacterized Rho GTPase-activating protein At5g61530 [Jatropha curcas] |
| 14 |
Hb_003728_020 |
0.0795957831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000035_380 |
0.080566987 |
- |
- |
- |
| 16 |
Hb_003005_020 |
0.0809289502 |
- |
- |
PREDICTED: DNA repair protein XRCC3 homolog [Jatropha curcas] |
| 17 |
Hb_000012_090 |
0.081233856 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 18 |
Hb_000365_360 |
0.0812527719 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000421_260 |
0.0814077049 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000019_230 |
0.0817777468 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |