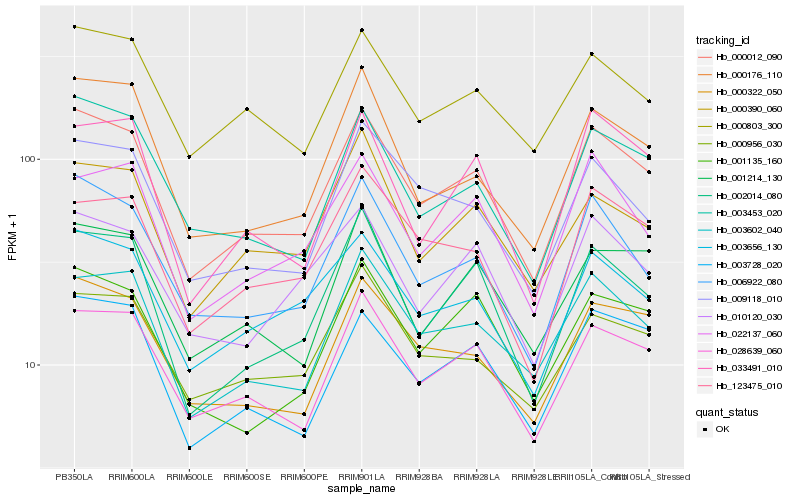
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_090 |
0.0 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 2 |
Hb_028639_060 |
0.0605555574 |
- |
- |
transporter, putative [Ricinus communis] |
| 3 |
Hb_006922_080 |
0.0635273267 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT3-like [Glycine max] |
| 4 |
Hb_003728_020 |
0.0661760543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003453_020 |
0.0667141132 |
- |
- |
PREDICTED: histidine biosynthesis bifunctional protein hisIE, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002014_080 |
0.0685424483 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 7 |
Hb_003602_040 |
0.068741136 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000322_050 |
0.0689708224 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 9 |
Hb_003656_130 |
0.0697135233 |
- |
- |
PREDICTED: protein bem46 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000176_110 |
0.0765839879 |
- |
- |
PREDICTED: serine--tRNA ligase [Jatropha curcas] |
| 11 |
Hb_000803_300 |
0.0775342005 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 12 |
Hb_009118_010 |
0.078667847 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 13 |
Hb_022137_060 |
0.0804521903 |
- |
- |
1-acylglycerophosphocholine O-acyltransferase [Vernicia fordii] |
| 14 |
Hb_001214_130 |
0.081233856 |
- |
- |
PREDICTED: uncharacterized protein LOC105646578 [Jatropha curcas] |
| 15 |
Hb_001135_160 |
0.0818068858 |
- |
- |
PREDICTED: elongator complex protein 6 [Jatropha curcas] |
| 16 |
Hb_010120_030 |
0.0827571429 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 17 |
Hb_123475_010 |
0.0829688702 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 18 |
Hb_000956_030 |
0.0836665516 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 19 |
Hb_033491_010 |
0.0844129558 |
- |
- |
Phosphatidylinositol N-acetylglucosaminyltransferase subunit C, putative [Ricinus communis] |
| 20 |
Hb_000390_060 |
0.0850152095 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g60570-like [Jatropha curcas] |