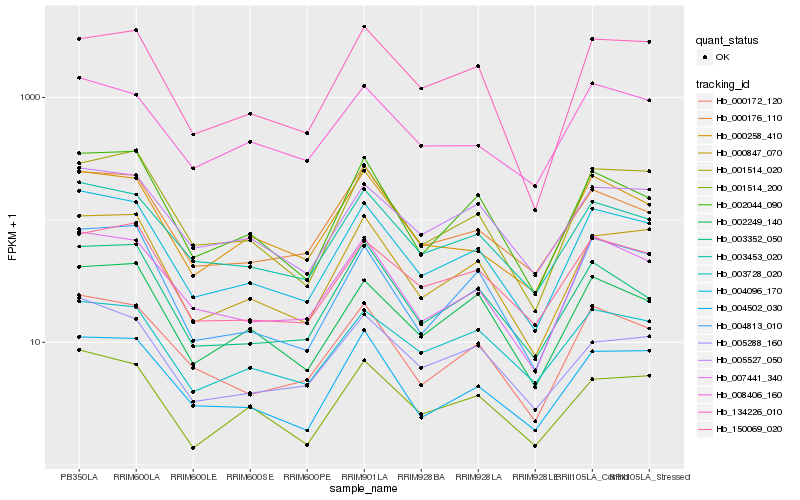
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0275933 [Jatropha curcas] |
| 2 |
Hb_000847_070 |
0.0600887142 |
- |
- |
PREDICTED: uncharacterized protein LOC105125699 [Populus euphratica] |
| 3 |
Hb_007441_340 |
0.0608208426 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_003453_020 |
0.0616763085 |
- |
- |
PREDICTED: histidine biosynthesis bifunctional protein hisIE, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000172_120 |
0.065247349 |
- |
- |
PREDICTED: uncharacterized protein LOC105650770 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002044_090 |
0.0739844121 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004813_010 |
0.078294301 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_004502_030 |
0.078643513 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_001514_020 |
0.0800321129 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_005527_050 |
0.0816205216 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 11 |
Hb_000258_410 |
0.0824842264 |
- |
- |
PREDICTED: uncharacterized protein LOC105630409 [Jatropha curcas] |
| 12 |
Hb_150069_020 |
0.0833773499 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000176_110 |
0.0839421941 |
- |
- |
PREDICTED: serine--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_002249_140 |
0.0843295004 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 15 |
Hb_008406_160 |
0.0865322336 |
- |
- |
PREDICTED: ADP,ATP carrier protein 3, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003728_020 |
0.0870998405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005288_160 |
0.0887129114 |
- |
- |
PREDICTED: uncharacterized protein LOC105647746 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001514_200 |
0.0896153358 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 19 |
Hb_134226_010 |
0.0906016964 |
- |
- |
40S ribosomal protein S21e, putative [Ricinus communis] |
| 20 |
Hb_003352_050 |
0.0908120857 |
- |
- |
PREDICTED: uncharacterized protein LOC105643709 [Jatropha curcas] |