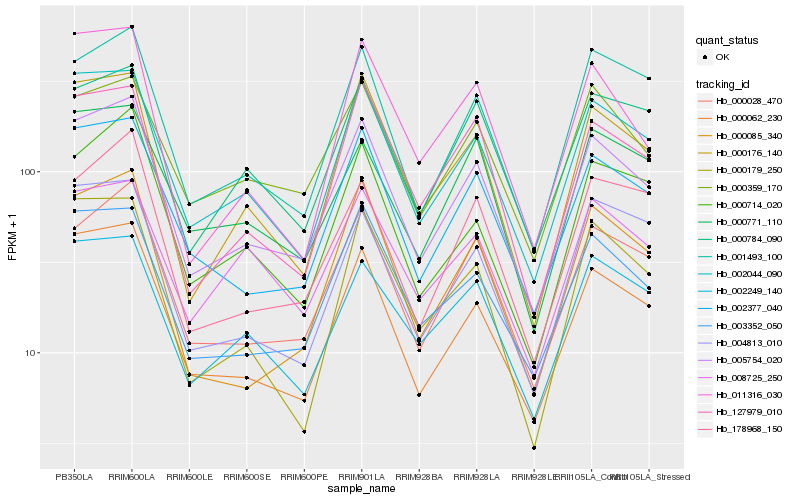
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005754_020 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF4 [Jatropha curcas] |
| 2 |
Hb_002044_090 |
0.0564904752 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000062_230 |
0.0568617617 |
- |
- |
PREDICTED: probable thimet oligopeptidase isoform X1 [Jatropha curcas] |
| 4 |
Hb_002377_040 |
0.0597227058 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000028_470 |
0.0609687667 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003352_050 |
0.0692875368 |
- |
- |
PREDICTED: uncharacterized protein LOC105643709 [Jatropha curcas] |
| 7 |
Hb_000176_140 |
0.0745387882 |
- |
- |
PREDICTED: bystin [Jatropha curcas] |
| 8 |
Hb_000085_340 |
0.078977068 |
- |
- |
PREDICTED: uncharacterized protein LOC105633038 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001493_100 |
0.0791444279 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 34-like [Jatropha curcas] |
| 10 |
Hb_000359_170 |
0.0812940942 |
- |
- |
hypothetical protein CISIN_1g014750mg [Citrus sinensis] |
| 11 |
Hb_002249_140 |
0.0841310295 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 12 |
Hb_000784_090 |
0.0860790975 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 13 |
Hb_127979_010 |
0.0868538537 |
- |
- |
PREDICTED: uncharacterized protein LOC105629383 [Jatropha curcas] |
| 14 |
Hb_000771_110 |
0.0921775071 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 15 |
Hb_000714_020 |
0.0923022851 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 16 |
Hb_000179_250 |
0.0924837618 |
- |
- |
PREDICTED: WD repeat-containing protein 61-like [Citrus sinensis] |
| 17 |
Hb_178968_150 |
0.0932109044 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_004813_010 |
0.0935877878 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_011316_030 |
0.0944000187 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008725_250 |
0.0963196786 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |