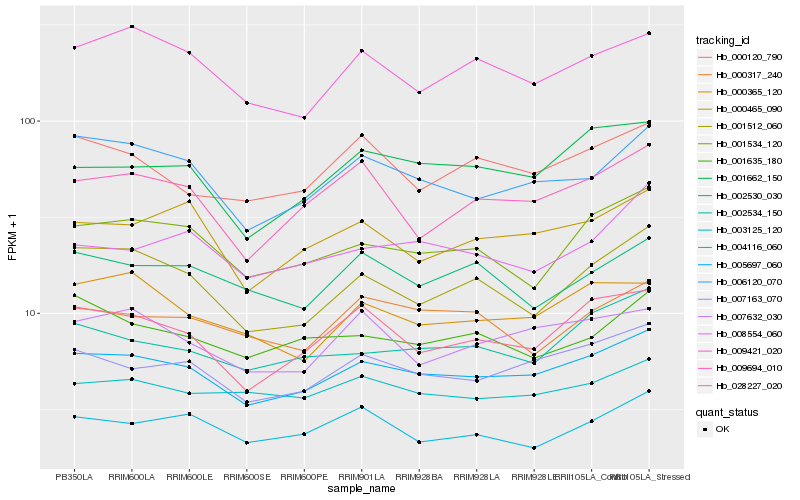
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005697_060 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 2 |
Hb_007163_070 |
0.0451247959 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006120_070 |
0.0612436341 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 4 |
Hb_028227_020 |
0.0633515543 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 5 |
Hb_000465_090 |
0.0676887324 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 6 |
Hb_001662_150 |
0.0676898846 |
- |
- |
Scaffold attachment factor B1 [Medicago truncatula] |
| 7 |
Hb_001534_120 |
0.0679002255 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 8 |
Hb_002534_150 |
0.0684305249 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_003125_120 |
0.0695680676 |
- |
- |
PREDICTED: TITAN-like protein [Jatropha curcas] |
| 10 |
Hb_007632_030 |
0.0718911016 |
- |
- |
PREDICTED: poly(A)-specific ribonuclease PARN isoform X1 [Jatropha curcas] |
| 11 |
Hb_000120_790 |
0.0727488027 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 12 |
Hb_001512_060 |
0.0731698374 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 13 |
Hb_009421_020 |
0.0735783129 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 14 |
Hb_009694_010 |
0.0740750946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000365_120 |
0.0763468359 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_008554_060 |
0.0773159431 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SPF27 homolog [Jatropha curcas] |
| 17 |
Hb_000317_240 |
0.0774638471 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 18 |
Hb_001635_180 |
0.0778608872 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At5g53840-like [Jatropha curcas] |
| 19 |
Hb_002530_030 |
0.0778966316 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 20 |
Hb_004116_060 |
0.0781997061 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |