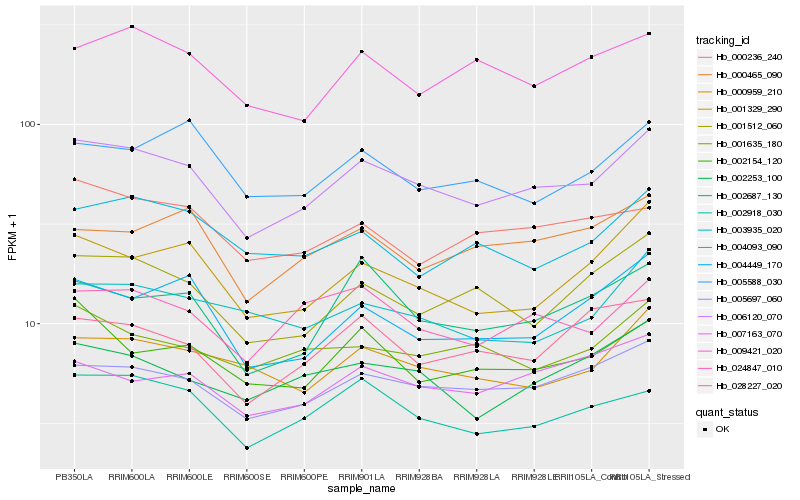
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_070 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 2 |
Hb_005697_060 |
0.0612436341 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_002918_030 |
0.0719830242 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 4 |
Hb_024847_010 |
0.076756252 |
- |
- |
UPF0586 C9orf41-like protein [Medicago truncatula] |
| 5 |
Hb_001512_060 |
0.0788384427 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000959_210 |
0.0801309659 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 7 |
Hb_001329_290 |
0.082261901 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 8 |
Hb_002253_100 |
0.0828954768 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004449_170 |
0.0831116188 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002687_130 |
0.0831496738 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001635_180 |
0.0841102471 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At5g53840-like [Jatropha curcas] |
| 12 |
Hb_009421_020 |
0.0855541998 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 13 |
Hb_002154_120 |
0.0856816152 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 14 |
Hb_007163_070 |
0.0858819485 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000236_240 |
0.0872432307 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 16 |
Hb_000465_090 |
0.0873069807 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 17 |
Hb_003935_020 |
0.0882003668 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_028227_020 |
0.0882306795 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 19 |
Hb_005588_030 |
0.0884657603 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 20 |
Hb_004093_090 |
0.0885952162 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |