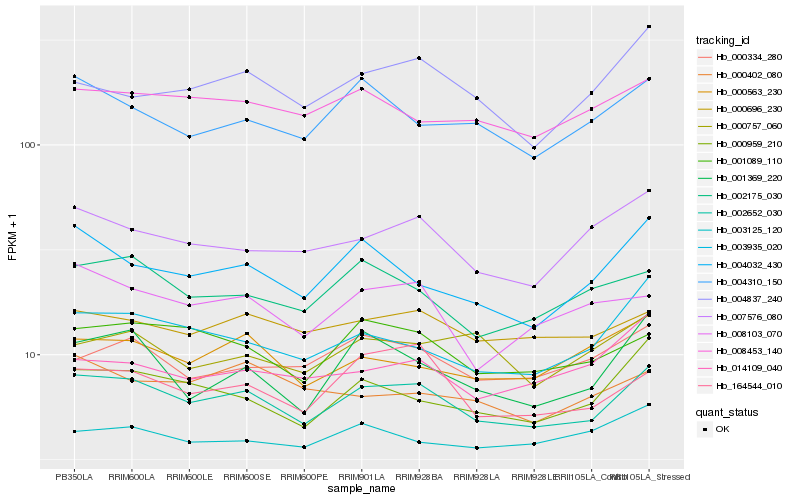
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002652_030 |
0.0 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 2 |
Hb_001089_110 |
0.0638318074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000696_230 |
0.0674373946 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 4 |
Hb_000959_210 |
0.0686993335 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 5 |
Hb_164544_010 |
0.072058297 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002175_030 |
0.0723100514 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 7 |
Hb_008453_140 |
0.0727465105 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 8 |
Hb_007576_080 |
0.0748899091 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 9 |
Hb_003935_020 |
0.0755761 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000334_280 |
0.075846945 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 11 |
Hb_000563_230 |
0.0765863036 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 12 |
Hb_004310_150 |
0.0771449322 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 13 |
Hb_004032_430 |
0.0805956743 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 14 |
Hb_008103_070 |
0.0811360472 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 15 |
Hb_003125_120 |
0.0811808833 |
- |
- |
PREDICTED: TITAN-like protein [Jatropha curcas] |
| 16 |
Hb_001369_220 |
0.0834566194 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_014109_040 |
0.0837567371 |
- |
- |
PREDICTED: probable folylpolyglutamate synthase [Jatropha curcas] |
| 18 |
Hb_000402_080 |
0.0852588977 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 19 |
Hb_000757_060 |
0.0858848943 |
- |
- |
hypothetical protein PRUPE_ppa001141mg [Prunus persica] |
| 20 |
Hb_004837_240 |
0.0859734856 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |