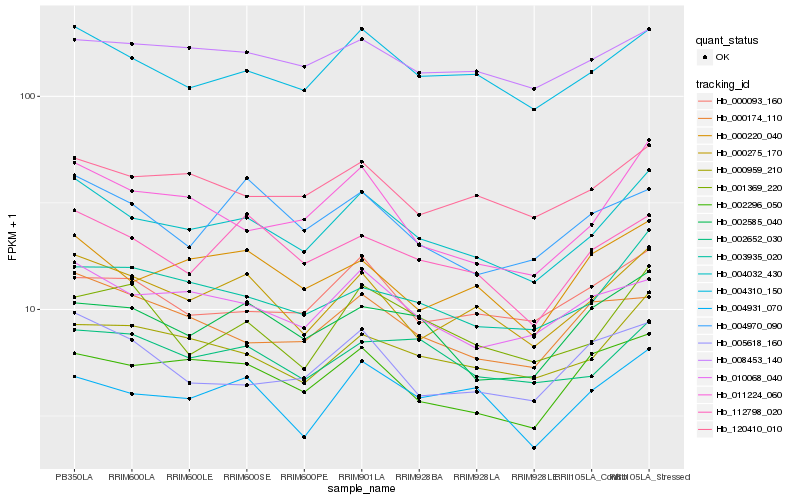
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_430 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 2 |
Hb_004310_150 |
0.0575967826 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 3 |
Hb_000275_170 |
0.0624544023 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 4 |
Hb_000959_210 |
0.0692421485 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 5 |
Hb_010068_040 |
0.070503378 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 6 |
Hb_011224_060 |
0.071536337 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 7 |
Hb_003935_020 |
0.0731779467 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_120410_010 |
0.0753399344 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 9 |
Hb_112798_020 |
0.0769839761 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 10 |
Hb_002296_050 |
0.0797135212 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 11 |
Hb_002652_030 |
0.0805956743 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 12 |
Hb_000220_040 |
0.0809043699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002585_040 |
0.0820743273 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 14 |
Hb_000093_160 |
0.08283804 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004931_070 |
0.0859467528 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000174_110 |
0.0876942799 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 17 |
Hb_008453_140 |
0.0877153036 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 18 |
Hb_001369_220 |
0.0883027956 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_005618_160 |
0.0884174615 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 20 |
Hb_004970_090 |
0.0887645427 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |