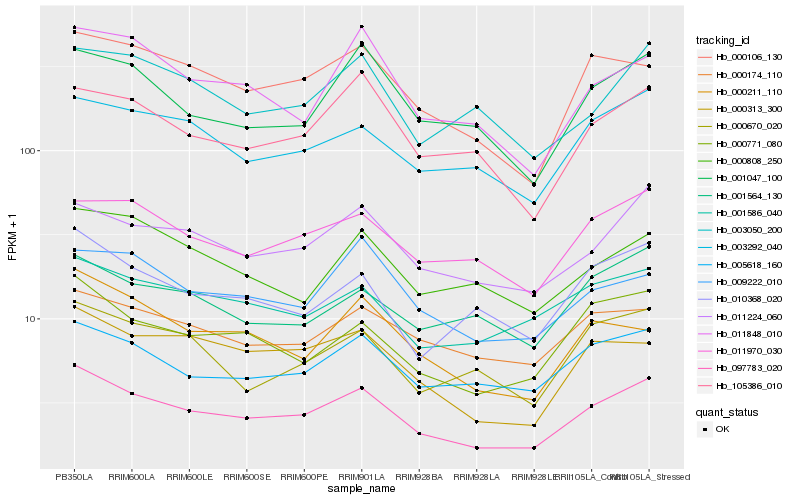
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097783_020 |
0.0 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 2 |
Hb_011224_060 |
0.0815239975 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 3 |
Hb_000771_080 |
0.0831149353 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 4 |
Hb_005618_160 |
0.085285491 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 5 |
Hb_003292_040 |
0.0904150821 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 6 |
Hb_011970_030 |
0.092469337 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 7 |
Hb_000313_300 |
0.0937509151 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Jatropha curcas] |
| 8 |
Hb_010368_020 |
0.09432865 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 9 |
Hb_000106_130 |
0.094535187 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001586_040 |
0.0950294786 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 11 |
Hb_001047_100 |
0.0960600459 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 12 |
Hb_105386_010 |
0.0977569278 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 13 |
Hb_000174_110 |
0.0982661538 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 14 |
Hb_000670_020 |
0.0999653956 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 15 |
Hb_003050_200 |
0.1026417015 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 16 |
Hb_011848_010 |
0.1028904872 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 17 |
Hb_001564_130 |
0.1041551549 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_009222_010 |
0.1044718361 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000211_110 |
0.1046592506 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_000808_250 |
0.1048847219 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |