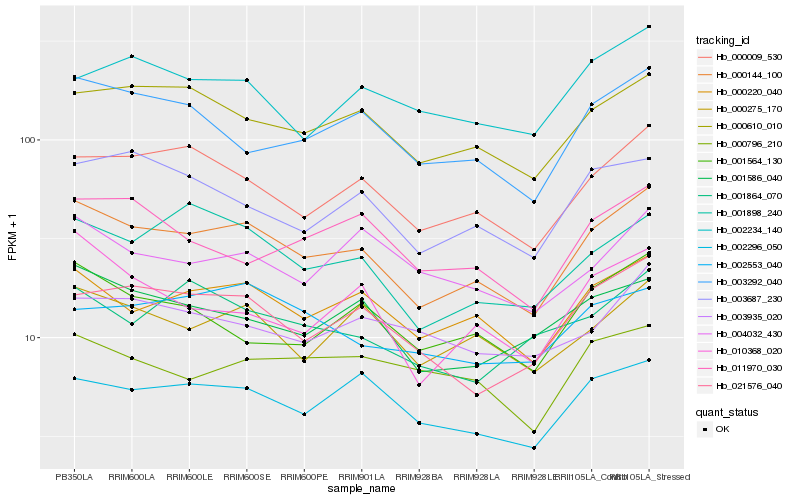
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000144_100 |
0.0 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000220_040 |
0.0689287227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000009_530 |
0.0748120841 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 4 |
Hb_000610_010 |
0.0785814882 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_000275_170 |
0.0829127766 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 6 |
Hb_002296_050 |
0.0854748742 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 7 |
Hb_003292_040 |
0.0861495337 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 8 |
Hb_021576_040 |
0.0868976635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001898_240 |
0.0884178381 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 10 |
Hb_001586_040 |
0.0905039695 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 11 |
Hb_003687_230 |
0.0922379956 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 12 |
Hb_001564_130 |
0.0948688915 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_010368_020 |
0.0991355845 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 14 |
Hb_003935_020 |
0.1018168261 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_011970_030 |
0.1021002069 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 16 |
Hb_002234_140 |
0.1022527095 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 17 |
Hb_004032_430 |
0.102911021 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 18 |
Hb_002553_040 |
0.1034366772 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 19 |
Hb_000796_210 |
0.1047773955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001864_070 |
0.1049111263 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |