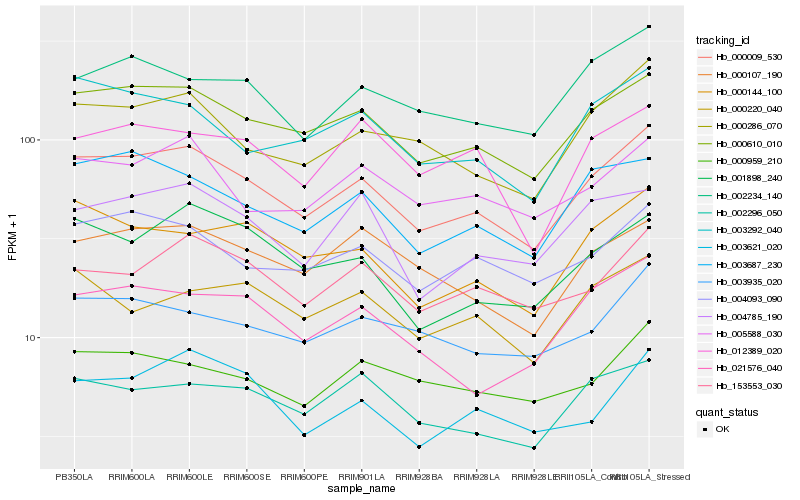
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_530 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 2 |
Hb_000610_010 |
0.0413382624 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 3 |
Hb_000286_070 |
0.06698475 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 4 |
Hb_003687_230 |
0.0712642175 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 5 |
Hb_004093_090 |
0.0719003286 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 6 |
Hb_000144_100 |
0.0748120841 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003292_040 |
0.0749775329 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 8 |
Hb_002234_140 |
0.0777021151 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 9 |
Hb_002296_050 |
0.0782984461 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 10 |
Hb_003935_020 |
0.0789681521 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_153553_030 |
0.0799133809 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 12 |
Hb_005588_030 |
0.0799162393 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 13 |
Hb_000107_190 |
0.0810025481 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 14 |
Hb_001898_240 |
0.0814112844 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 15 |
Hb_021576_040 |
0.0821144415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004785_190 |
0.082374631 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 17 |
Hb_000959_210 |
0.0837674159 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 18 |
Hb_003621_020 |
0.0859105068 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 19 |
Hb_000220_040 |
0.0867897698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012389_020 |
0.0893436808 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |