| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
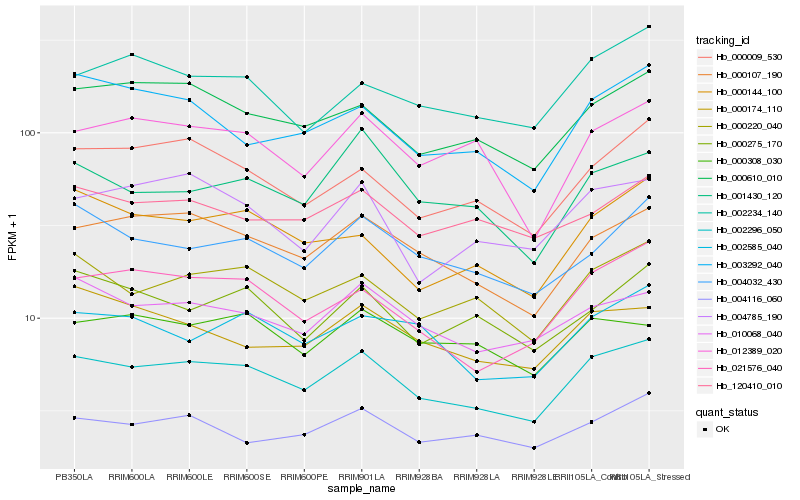
Hb_002296_050 |
0.0 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 2 |
Hb_000107_190 |
0.0609323732 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 3 |
Hb_000220_040 |
0.0663400299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000610_010 |
0.0698028699 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_010068_040 |
0.0739169722 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 6 |
Hb_021576_040 |
0.0761331359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000009_530 |
0.0782984461 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 8 |
Hb_004032_430 |
0.0797135212 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 9 |
Hb_002585_040 |
0.080098577 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 10 |
Hb_012389_020 |
0.0828813694 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 11 |
Hb_004116_060 |
0.0837854412 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 12 |
Hb_000144_100 |
0.0854748742 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001430_120 |
0.0857207598 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 14 |
Hb_004785_190 |
0.085839291 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 15 |
Hb_120410_010 |
0.0859035974 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 16 |
Hb_002234_140 |
0.0863301382 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 17 |
Hb_000275_170 |
0.086417602 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 18 |
Hb_003292_040 |
0.0877836892 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 19 |
Hb_000308_030 |
0.0880379478 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 20 |
Hb_000174_110 |
0.0884401522 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |