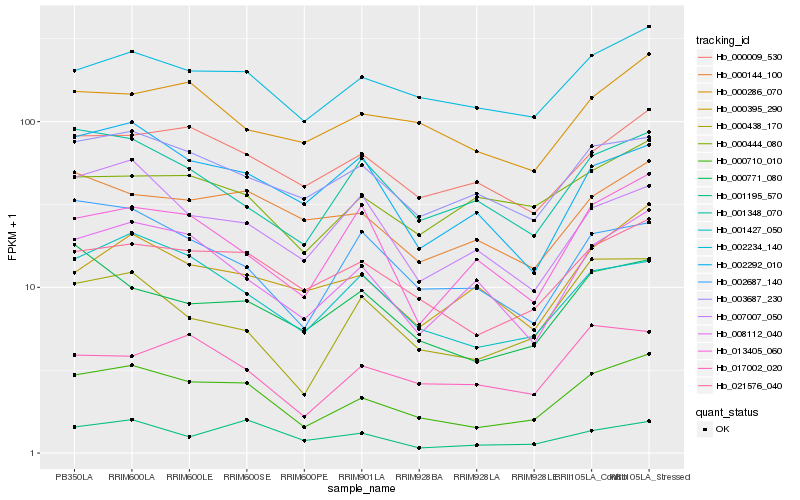
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000710_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630356 [Jatropha curcas] |
| 2 |
Hb_021576_040 |
0.0902232113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002234_140 |
0.1095998983 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 4 |
Hb_001427_050 |
0.110878979 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 5 |
Hb_008112_040 |
0.1133899142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 6 |
Hb_003687_230 |
0.1186295517 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 7 |
Hb_000438_170 |
0.1199178852 |
- |
- |
- |
| 8 |
Hb_000009_530 |
0.121457743 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 9 |
Hb_001195_570 |
0.1258461555 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 10 |
Hb_000144_100 |
0.1276744157 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000771_080 |
0.1283245498 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 12 |
Hb_001348_070 |
0.1293241485 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_000286_070 |
0.129365528 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 14 |
Hb_017002_020 |
0.1310551728 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 15 |
Hb_002687_140 |
0.1311356575 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 16 |
Hb_000395_290 |
0.1317033667 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 17 |
Hb_013405_060 |
0.1333343518 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000444_080 |
0.1353748568 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 19 |
Hb_007007_050 |
0.1364594574 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 20 |
Hb_002292_010 |
0.1366371888 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |