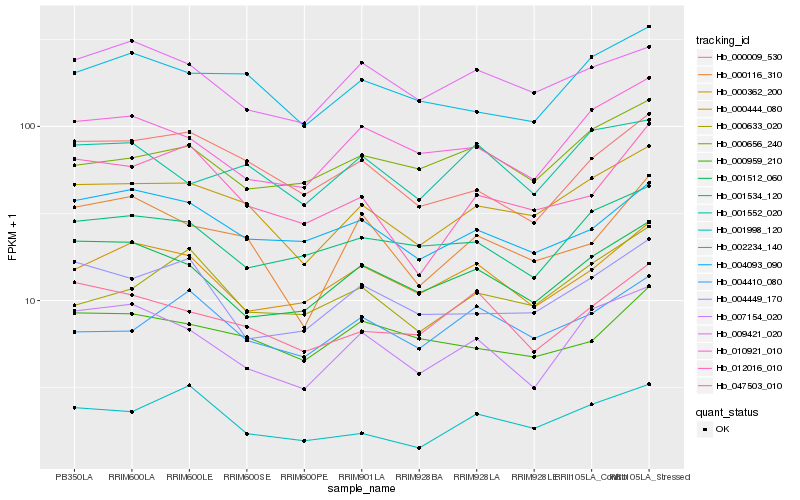
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000444_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 2 |
Hb_002234_140 |
0.0755854846 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 3 |
Hb_010921_010 |
0.0914751167 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 4 |
Hb_000009_530 |
0.0916585422 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 5 |
Hb_001552_020 |
0.0938468491 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 6 |
Hb_000116_310 |
0.0953183932 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |
| 7 |
Hb_001512_060 |
0.0957201176 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001534_120 |
0.0960022131 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 9 |
Hb_047503_010 |
0.0971349208 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 10 |
Hb_004093_090 |
0.0976092053 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 11 |
Hb_004449_170 |
0.0985353487 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000959_210 |
0.098826483 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 13 |
Hb_007154_020 |
0.0990025471 |
- |
- |
- |
| 14 |
Hb_009421_020 |
0.0991672013 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 15 |
Hb_004410_080 |
0.09981521 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000362_200 |
0.100743769 |
- |
- |
exonuclease family protein [Populus trichocarpa] |
| 17 |
Hb_000656_240 |
0.102013721 |
- |
- |
proteasome subunit alpha type, putative [Ricinus communis] |
| 18 |
Hb_000633_020 |
0.1022368007 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 19 |
Hb_012016_010 |
0.1025219503 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001998_120 |
0.1025738468 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |