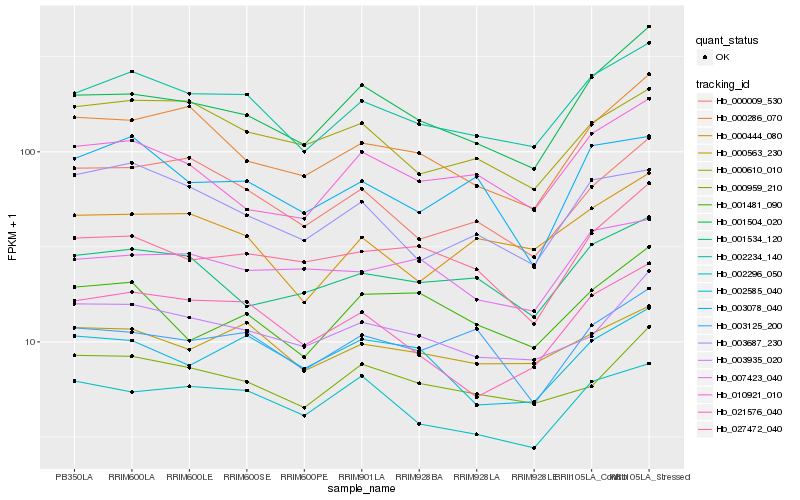
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002234_140 |
0.0 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 2 |
Hb_001504_020 |
0.0728389177 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 3 |
Hb_021576_040 |
0.0741348133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000444_080 |
0.0755854846 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 5 |
Hb_000563_230 |
0.0762706751 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 6 |
Hb_003935_020 |
0.0774315442 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000009_530 |
0.0777021151 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 8 |
Hb_000959_210 |
0.079480835 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 9 |
Hb_000286_070 |
0.0799203933 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 10 |
Hb_001534_120 |
0.0828553551 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 11 |
Hb_010921_010 |
0.0854240461 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 12 |
Hb_000610_010 |
0.0854656472 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_002296_050 |
0.0863301382 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 14 |
Hb_003687_230 |
0.0864791116 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 15 |
Hb_027472_040 |
0.0866140448 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 16 |
Hb_003125_200 |
0.0884659206 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 17 |
Hb_003078_040 |
0.0886478423 |
- |
- |
PREDICTED: hit family protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007423_040 |
0.0889125493 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002585_040 |
0.0911103118 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 20 |
Hb_001481_090 |
0.0938216666 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |